













| General Information: |
|
| Name(s) found: |
RAD54 /
YGL163C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 898 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IMP |
| Biological Process: |
chromatin remodeling
[IDA heteroduplex formation [IDA [NOT]double-strand break repair via single-strand annealing [IMP telomere maintenance via recombination [IMP double-strand break repair via synthesis-dependent strand annealing [TAS |
| Molecular Function: |
DNA-dependent ATPase activity
[IDA DNA topoisomerase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARRRLPDRP PNGIGAGERP RLVPRPINVQ DSVNRLTKPF RVPYKNTHIP PAAGRIATGS 60
61 DNIVGGRSLR KRSATVCYSG LDINADEAEY NSQDISFSQL TKRRKDALSA QRLAKDPTRL 120
121 SHIQYTLRRS FTVPIKGYVQ RHSLPLTLGM KKKITPEPRP LHDPTDEFAI VLYDPSVDGE 180
181 MIVHDTSMDN KEEESKKMIK STQEKDNINK EKNSQEERPT QRIGRHPALM TNGVRNKPLR 240
241 ELLGDSENSA ENKKKFASVP VVIDPKLAKI LRPHQVEGVR FLYRCVTGLV MKDYLEAEAF 300
301 NTSSEDPLKS DEKALTESQK TEQNNRGAYG CIMADEMGLG KTLQCIALMW TLLRQGPQGK 360
361 RLIDKCIIVC PSSLVNNWAN ELIKWLGPNT LTPLAVDGKK SSMGGGNTTV SQAIHAWAQA 420
421 QGRNIVKPVL IISYETLRRN VDQLKNCNVG LMLADEGHRL KNGDSLTFTA LDSISCPRRV 480
481 ILSGTPIQND LSEYFALLSF SNPGLLGSRA EFRKNFENPI LRGRDADATD KEITKGEAQL 540
541 QKLSTIVSKF IIRRTNDILA KYLPCKYEHV IFVNLKPLQN ELYNKLIKSR EVKKVVKGVG 600
601 GSQPLRAIGI LKKLCNHPNL LNFEDEFDDE DDLELPDDYN MPGSKARDVQ TKYSAKFSIL 660
661 ERFLHKIKTE SDDKIVLISN YTQTLDLIEK MCRYKHYSAV RLDGTMSINK RQKLVDRFND 720
721 PEGQEFIFLL SSKAGGCGIN LIGANRLILM DPDWNPAADQ QALARVWRDG QKKDCFIYRF 780
781 ISTGTIEEKI FQRQSMKMSL SSCVVDAKED VERLFSSDNL RQLFQKNENT ICETHETYHC 840
841 KRCNAQGKQL KRAPAMLYGD ATTWNHLNHD ALEKTNDHLL KNEHHYNDIS FAFQYISH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
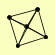
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
RAD51 RAD52 RAD54 RAD55 RAD57 |
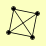
| View Details | Ho Y, et al. (2002) |
|
MDH1 MGE1 RAD54 TMA19 |
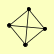
| View Details | Qiu et al. (2008) |
|
RAD51 RAD54 RAD55 RAD57 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
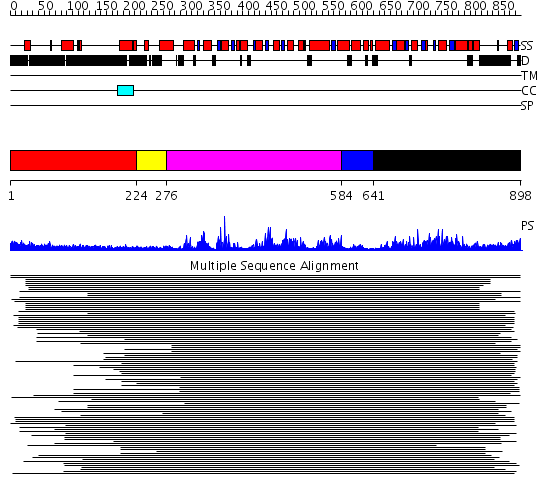
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..223] | 1.238994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [224..275] | 13.09 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 3 | View Details | [276..583] | 13.09 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [584..640] | 8.221849 | Putative DEAD box RNA helicase | |
| 5 | View Details | [641..898] | 7.045757 | Nucleotide excision repair enzyme UvrB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)