













| General Information: |
|
| Name(s) found: |
PAN2 /
YGL094C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1115 amino acids |
Gene Ontology: |
|
| Cellular Component: |
PAN complex
[IDA cytoplasm [IDA |
| Biological Process: |
mRNA 3'-end processing
[IMP mRNA processing [IMP postreplication repair [IGI |
| Molecular Function: |
poly(A)-specific ribonuclease activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNWQHFFNN PVDLSEHLKK PYFRFDNRDK EITAISFDEK ANLIWSGDSY GCISSYDPTF 60
61 QLYTRYRGHI GGNSVKDILS HRDGILSISE DSLHFANRRG VTKLNLTSID IAAFSELNTM 120
121 CYSPHSLKNN IYCGGDNTNW GIASIDLNRG CLDSLLNYSS KVKLMCSNNK VLSIGRQTGT 180
181 VDLLDPTSNR TIKSFNAHSA SISAMDLRDN TLVTVGKSKR FYNLYADPFV NVYDLRTMRQ 240
241 LPPVSFSKGT TMGSGGADFV QLHPLLPTVM IVASSSGSFD FIDLSNPTLR TQYVHPCQSI 300
301 KKLCLSPNGD VLGILEADNH LDTWRRSSNN MGMFTNTPEM LAYPDYFNDI TSDGPISVDD 360
361 ETYPLSSVGM PYYLDKLLSA WPPVVFKSEG TIPQLTGKSP LPSSGKLKSN LAVISSQNEK 420
421 LSTQEFPLLR YDRTKYGMRN AIPDYVCLRD IRKQITSGLE TSDIQTYTSI NKYEVPPAYS 480
481 RLPLTSGRFG TDNFDFTPFN NTEYSGLDPD VDNHYTNAII QLYRFIPEMF NFVVGCLKDE 540
541 NFETTLLTDL GYLFDMMERS HGKICSSSNF QASLKSLTDK RQLENGEPQE HLEEYLESLC 600
601 IRESIEDFNS SESIKRNMPQ KFNRFLLSQL IKEEAQTVNH NITLNQCFGL ETEIRTECSC 660
661 DHYDTTVKLL PSLSISGINK TVIKQLNKKS NGQNILPYIE YAMKNVTQKN SICPTCGKTE 720
721 TITQECTVKN LPSVLSLELS LLDTEFSNIR SSKNWLTSEF YGSIIKNKAV LRSTASELKG 780
781 TSHIFKYELN GYVAKITDNN NETRLVTYVK KYNPKENCFK WLMFNDYLVV EITEEEALKM 840
841 TYPWKTPEII IYCDAEELRK PFFSVDTYSI NYDILFRDYF ANGIRDTARR EYKLLTHDEA 900
901 PKSGTLVAID AEFVSLQSEL CEIDHQGIRS IIRPKRTALA RISIIRGEEG ELYGVPFVDD 960
961 YVVNTNHIED YLTRYSGILP GDLDPEKSTK RLVRRNVVYR KVWLLMQLGC VFVGHGLNND1020
1021 FKHININVPR NQIRDTAIYF LQGKRYLSLR YLAYVLLGMN IQEGNHDSIE DAHTALILYK1080
1081 KYLHLKEKAI FEKVLNSVYE EGRAHNFKVP ETSKG |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
PAN2 PAN3 |
| View Details | Krogan NJ, et al. (2006) |

|
PAN2 PAN3 |
| View Details | Gavin AC, et al. (2006) |
|
PAB1 PAN2 PAN3 |
| View Details | Gavin AC, et al. (2002) |

|
PAN2 PAN3 |
| View Details | Qiu et al. (2008) |
|
PAN2 PAN3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
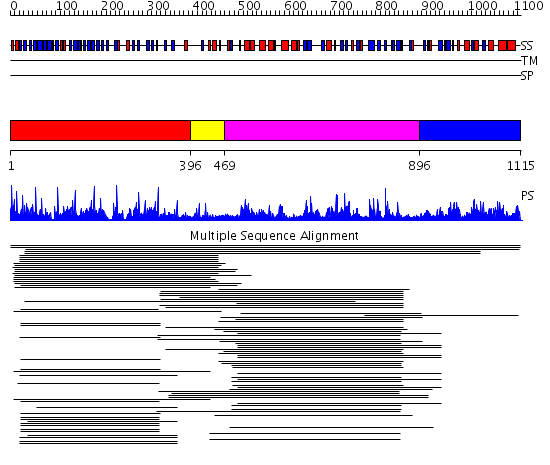
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..395] | 75.522879 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 2 | View Details | [396..468] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [469..895] | 58.218995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [896..1115] | 7.52 | Exonuclease I |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)