













| General Information: |
|
| Name(s) found: |
GEA2 /
YEL022W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1459 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi cis cisterna
[IDA Golgi-associated vesicle [TAS |
| Biological Process: |
ER to Golgi vesicle-mediated transport
[TAS retrograde vesicle-mediated transport, Golgi to ER [IGI intra-Golgi vesicle-mediated transport [TAS actin cytoskeleton organization [IGI |
| Molecular Function: |
ARF guanyl-nucleotide exchange factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDREFVTVD PVTIIIKECI NLSTAMRKYS KFTSQSGVAA LLGGGSEIFS NQDDYLAHTF 60
61 NNLNTNKHND PFLSGFIQLR LMLNKLKNLD NIDSLTILQP FLLIVSTSSI SGYITSLALD 120
121 SLQKFFTLNI INESSQNYIG AHRATVNALT HCRFEGSQQL SDDSVLLKVV FLLRSIVDSP 180
181 YGDLLSNSII YDVLQTILSL ACNNRRSEVL RNAAQSTMIA VTVKIFSKLK TIEPVNVNQI 240
241 YINDESYTND VLKADTIGTN VESKEEGSQE DPIGMKVNNE EAISEDDGIE EEHIHSEKST 300
301 NGAEQLDIVQ KTTRSNSRIQ AYADDNYGLP VVRQYLNLLL SLIAPENELK HSYSTRIFGL 360
361 ELIQTALEIS GDRLQLYPRL FTLISDPIFK SILFIIQNTT KLSLLQATLQ LFTTLVVILG 420
421 NNLQLQIELT LTRIFSILLD DGTANNSSSE NKNKPSIIKE LLIEQISILW TRSPSFFTST 480
481 FINFDCNLDR ADVSINFLKA LTKLALPESA LTTTESVPPI CLEGLVSLVD DMFDHMKDID 540
541 REEFGRQKNE MEILKKRDRK TEFIECTNAF NEKPKKGIPM LIEKGFIASD SDKDIAEFLF 600
601 NNNNRMNKKT IGLLLCHPDK VSLLNEYIRL FDFSGLRVDE AIRILLTKFR LPGESQQIER 660
661 IIEAFSSAYC ENQDYDPSKI SDNAEDDIST VQPDADSVFI LSYSIIMLNT DLHNPQVKEH 720
721 MSFEDYSGNL KGCCNHKDFP FWYLDRIYCS IRDKEIVMPE EHHGNEKWFE DAWNNLISST 780
781 TVITEIKKDT QSVMDKLTPL ELLNFDRAIF KQVGPSIVST LFNIYVVASD DHISTRMITS 840
841 LDKCSYISAF FDFKDLFNDI LNSIAKGTTL INSSHDDELS TLAFEYGPMP LVQIKFEDTN 900
901 TEIPVSTDAV RFGRSFKGQL NTVVFFRIIR RNKDPKIFSK ELWLNIVNII LTLYEDLILS 960
961 PDIFPDLQKR LKLSNLPKPS PEISINKSKE SKGLLSTFAS YLKGDEEPTE EEIKSSKKAM1020
1021 ECIKSSNIAA SVFGNESNIT ADLIKTLLDS AKTEKNADNS RYFEAELLFI IELTIALFLF1080
1081 CKEEKELGKF ILQKVFQLSH TKGLTKRTVR RMLTYKILLI SLCADQTEYL SKLINDELLK1140
1141 KGDIFTQKFF ATNQGKEFLK RLFSLTESEF YRGFLLGNEN FWKFLRKVTA MKEQSESIFE1200
1201 YLNESIKTDS NILTNENFMW VLGLLDEISS MGAVGNHWEI EYKKLTESGH KIDKENPYKK1260
1261 SIELSLKSIQ LTSHLLEDNN DLRKNEIFAI IQALAHQCIN PCKQISEFAV VTLEQTLINK1320
1321 IEIPTNEMES VEELIEGGLL PLLNSSETQE DQKILISSIL TIISNVYLHY LKLGKTSNET1380
1381 FLKILSIFNK FVEDSDIEKK LQQLILDKKS IEKGNGSSSH GSAHEQTPES NDVEIEATAP1440
1441 IDDNTDDDNK PKLSDVEKD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CLU1 CRM1 FKS1 GEA2 KAP104 MIR1 YHR020W |
| View Details | Gavin AC, et al. (2002) |
|
ADH1 BMH1 BMH2 CLU1 CRM1 ECM29 FET5 FKS1 FTH1 GCN1 GEA2 GFA1 HRP1 KAP104 KAP123 LCB1 LCB2 LYS12 MIR1 NAB2 RPN3 SAM1 SAM2 YHR020W |
| View Details | Qiu et al. (2008) |
|
GEA1 GEA2 GLO3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
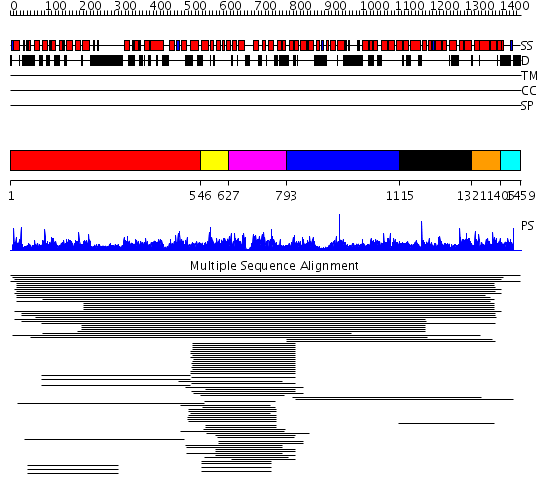
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..545] | 1.046927 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [546..626] | 1294.154902 | ARF guanine-exchange factor 2, Gea2 | |
| 3 | View Details | [627..792] | 1294.154902 | ARF guanine-exchange factor 2, Gea2 | |
| 4 | View Details | [793..1114] | 9.55 | Importin beta | |
| 5 | View Details | [1115..1320] | 9.55 | Importin beta | |
| 6 | View Details | [1321..1404] | 9.55 | Importin beta | |
| 7 | View Details | [1405..1459] | 7.21 | Serum albumin | |
| 8 | View Details | [1177..1252] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1253..1459] | 3.112988 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)