













| General Information: |
|
| Name(s) found: |
EGD1 /
YPL037C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 157 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nascent polypeptide-associated complex
[IDA |
| Biological Process: |
'de novo' cotranslational protein folding
[IPI |
| Molecular Function: |
unfolded protein binding
[IMP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
EGD1 EGD2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
EGD1 EGD2 |
| View Details | Krogan NJ, et al. (2006) |

|
BTT1 CAF130 EGD1 EGD2 PLC1 YJR011C |
| View Details | Ho Y, et al. (2002) |
|
AAT2 ACH1 ACO1 AFR1 AHP1 ARC1 ARF1 ATP3 CAR2 CCT5 CDC33 CKA1 CKB1 CKB2 CLU1 COF1 COR1 CPR1 CYR1 CYS4 DBP2 DUN1 EGD1 ENP2 ERB1 ERG13 ERG6 FPR1 FPR3 FRS2 GCD11 GND1 GND2 GRX1 GUS1 HAS1 HEF3 HHF1, HHF2 HHT1, HHT2 HOT1 HTA1 HTB2 KRE33 KRI1 LRO1 LSP1 MET6 MGM101 NAP1 NMD3 NOG1 NOP12 NOP2 NOP4 NOP56 NTF2 NUG1 OLA1 OYE2 PDI1 PFK1 PIL1 POB3 PRM2 PRP19 PUF6 PWP1 QCR2 RNR2 RSN1 SAH1 SCP160 SEC53 SEN15 SES1 SNU13 SOD1 SPT16 SSF1 TCP1 TEF4 THS1 TIF1, TIF2 TIF6 TMA19 TSA2 UBA1 VMA4 VMA5 WTM1 YER084W YJU2 YNK1 |
| View Details | Qiu et al. (2008) |

|
BTT1 EGD1 EGD2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
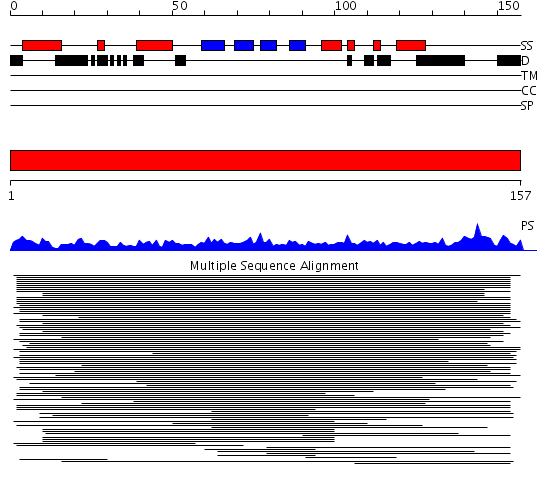
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 27.920819 | NAC domain No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)