













| General Information: |
|
| Name(s) found: |
NUP2 /
YLR335W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 720 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromatin
[IDA mitochondrion [IDA nuclear pore [IDA |
| Biological Process: |
mRNA export from nucleus
[TAS rRNA export from nucleus [TAS nuclear pore organization [TAS snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS chromatin silencing [IMP protein export from nucleus [TAS chromatin silencing at telomere [IMP ribosomal protein import into nucleus [TAS |
| Molecular Function: |
structural molecule activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRVADAQI QRETYDSNES DDDVTPSTKV ASSAVMNRRK IAMPKRRMAF KPFGSAKSDE 60
61 TKQASSFSFL NRADGTGEAQ VDNSPTTESN SRLKALNLQF KAKVDDLVLG KPLADLRPLF 120
121 TRYELYIKNI LEAPVKSIEN PTQTKGNDAK PAKVEDVQKS SDSSSEDEVK VEGPKFTIDA 180
181 KPPISDSVFS FGPKKENRKK DESDSENDIE IKGPEFKFSG TVSSDVFKLN PSTDKNEKKT 240
241 ETNAKPFSFS SATSTTEQTK SKNPLSLTEA TKTNVDNNSK AEASFTFGTK HAADSQNNKP 300
301 SFVFGQAAAK PSLEKSSFTF GSTTIEKKND ENSTSNSKPE KSSDSNDSNP SFSFSIPSKN 360
361 TPDASKPSFS FGVPNSSKNE TSKPVFSFGA ATPSAKEASQ EDDNNNVEKP SSKPAFNLIS 420
421 NAGTEKEKES KKDSKPAFSF GISNGSESKD SDKPSLPSAV DGENDKKEAT KPAFSFGINT 480
481 NTTKTADTKA PTFTFGSSAL ADNKEDVKKP FSFGTSQPNN TPSFSFGKTT ANLPANSSTS 540
541 PAPSIPSTGF KFSLPFEQKG SQTTTNDSKE ESTTEATGNE SQDATKVDAT PEESKPINLQ 600
601 NGEEDEVALF SQKAKLMTFN AETKSYDSRG VGEMKLLKKK DDPSKVRLLC RSDGMGNVLL 660
661 NATVVDSFKY EPLAPGNDNL IKAPTVAADG KLVTYIVKFK QKEEGRSFTK AIEDAKKEMK 720
721 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
KAP95 NUP2 NUP60 SRP1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
GLE2 NOP14 NUP1 NUP100 NUP116 NUP133 NUP157 NUP159 NUP170 NUP188 NUP2 NUP42 POM152 |
| View Details | Krogan NJ, et al. (2006) |
|
CBC2 KAP95 NUP1 NUP2 NUP60 OGG1 PCT1 RAD53 SRP1 STO1 ULP1 URB2 YRA2 |
| View Details | Gavin AC, et al. (2006) |
|
KAP95 NUP2 NUP60 SRP1 |
| View Details | Ho Y, et al. (2002) |

|
DIS3 EAF3 FIP1 HAS1 HPR1 KAP95 MES1 MFT1 NAM8 NOP6 NUP1 NUP2 NUP60 PAP1 PCT1 REB1 RNT1 RRP4 RRP43 RRP6 RTT103 SIF2 SIN3 SNU56 SRP1 STO1 THO2 TRA1 UME1 YPR090W |
| View Details | Qiu et al. (2008) |

|
ASM4 GLE2 KAP120 MEX67 NDC1 NIC96 NSP1 NUP1 NUP100 NUP116 NUP133 NUP145 NUP157 NUP159 NUP170 NUP188 NUP2 NUP42 NUP49 NUP57 NUP82 NUP85 POM152 SEC13 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
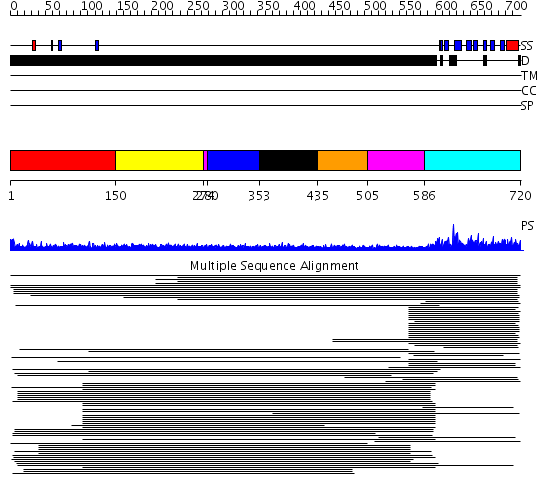
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [150..273] | 14.0 | Sec24 | |
| 3 | View Details | [274..279] [505..585] |
11.01 | No description for 1l8mA was found. | |
| 4 | View Details | [280..352] | 11.01 | No description for 1l8mA was found. | |
| 5 | View Details | [353..434] | 11.01 | No description for 1l8mA was found. | |
| 6 | View Details | [435..504] | 11.01 | No description for 1l8mA was found. | |
| 7 | View Details | [586..720] | 134.190949 | Nuclear pore complex protein Nup358 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)