













| General Information: |
|
| Name(s) found: |
NUP133 /
YKR082W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1157 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[IDA |
| Biological Process: |
mRNA export from nucleus
[IMP rRNA export from nucleus [TAS nuclear pore organization [IMP snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS chromosome localization [IMP tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS DNA metabolic process [IGI protein export from nucleus [TAS ribosomal protein import into nucleus [TAS |
| Molecular Function: |
structural molecule activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEKKVHLRL RKELSVPIAV VENESLAQLS YEEESQASLM DISMEQQQLR LHSHFDNSKV 60
61 FTENNRYIVK TLQTDYSSGF SNDDELNGYI DMQIGYGLVN DHKKVYIWNI HSTQKDTPYI 120
121 TVPFRSDDND EIAVAPRCIL TFPATMDESP LALNPNDQDE TGGLIIIKGS KAIYYEDINS 180
181 INNLNFKLSE KFSHELELPI NSSGGEKCDL MLNCEPAGIV LSTNMGRIFF ITIRNSMGKP 240
241 QLKLGKLLNK PFKLGIWSKI FNTNSSVVSL RNGPILGKGT RLVYITTNKG IFQTWQLSAT 300
301 NSHPTKLIDV NIYEAILESL QDLYPFAHGT LKIWDSHPLQ DESSQLFLSS IYDSSCNETY 360
361 YILSTIIFDS SSNSFTIFST YRLNTFMESI TDTKFKPKIF IPQMENANDT NEVTSILVMF 420
421 PNAVVITQVN SKLDSSYSMR RKWEDIVSLR NDIDIIGSGY DSKSLYVLTK QMGVLQFFVK 480
481 ENEETNSKPE VGFVKSHVDQ AVYFSKINAN PIDFNLPPEI SLDQESIEHD LKLTSEEIFH 540
541 SNGKYIPPML NTLGQHLSVR KEFFQNFLTF VAKNFNYKIS PELKLDLIEK FEILNCCIKF 600
601 NSIIRQSDVL NDIWEKTLSN YNLTQNEHLT TKTVVINSPD VFPVIFKQFL NHVVFVLFPS 660
661 QNQNFKLNVT NLINLCFYDG ILEEGEKTIR YELLELDPME VDTSKLPWFI NFDYLNCINQ 720
721 CFFDFTFACE EEGSLDSYKE GLLKIVKILY YQFNQFKIWI NTQPVKSVNA NDNFININNL 780
781 YDDNHLDWNH VLCKVNLKEQ CIQIAEFYKD LSGLVQTLQT LDQNDSTTVS LYETFFNEFP 840
841 KEFSFTLFEY LIKHKKLNDL IFRFPQQHDV LIQFFQESAP KYGHVAWIQQ ILDGSYADAM 900
901 NTLKNITVDD SKKGESLSEC ELHLNVAKLS SLLVEKDNLD INTLRKIQYN LDTIDAEKNI 960
961 SNKLKKGEVQ ICKRFKNGSI REVFNILVEE LKSTTVVNLS DLVELYSMLD DEESLFIPLR1020
1021 LLSVDGNLLN FEVKKFLNAL VWRRIVLLNA SNEGDKLLQH IVKRVFDEEL PKNNDFPLPS1080
1081 VDLLCDKSLL TPEYISETYG RFPIDQNAIR EEIYEEISQV ETLNSDNSLE IKLHSTIGSV1140
1141 AKEKNYTINY ETNTVEY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
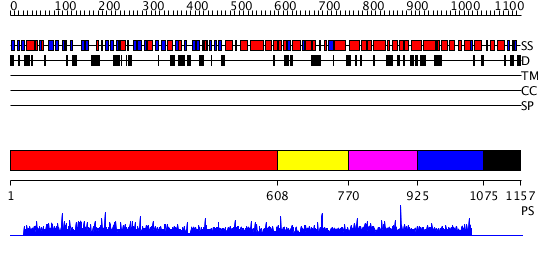
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..607] | 6.44 | The crystal structure of the N-terminal domain of Nup133 reveals a beta-propeller fold common to several nucleoporins | |
| 2 | View Details | [608..769] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [770..924] | 2.042996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [925..1074] | 1.040977 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1075..1157] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)