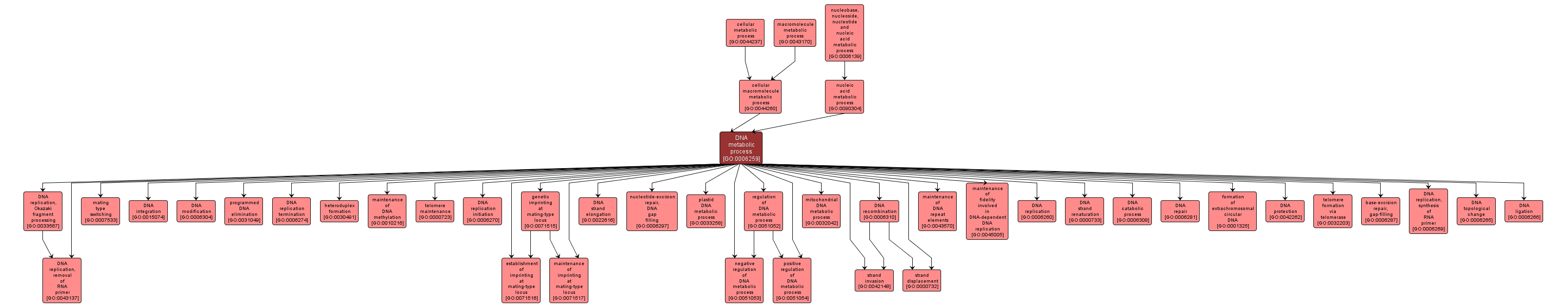
GO TERM SUMMARY
|
| Name: |
DNA metabolic process |
| Acc: |
GO:0006259 |
| Aspect: |
Biological Process |
| Desc: |
Any cellular metabolic process involving deoxyribonucleic acid. This is one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides. |
Synonyms:
- GO:0055132
- DNA metabolism
- cellular DNA metabolism
|
|

|
INTERACTIVE GO GRAPH
|