













| General Information: |
|
| Name(s) found: |
NUP84 /
YDL116W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 726 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[IDA |
| Biological Process: |
DNA metabolic process
[IGI protein export from nucleus [TAS response to DNA damage stimulus [IMP ribosomal protein import into nucleus [TAS mRNA export from nucleus [IMP rRNA export from nucleus [TAS nuclear envelope organization [IMP nuclear pore organization [IMP snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS chromosome localization [IMP tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS |
| Molecular Function: |
structural molecule activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELSPTYQTE RFTKFSDTLK EFKIEQNNEQ NPIDPFNIIR EFRSAAGQLA LDLANSGDES 60
61 NVISSKDWEL EARFWHLVEL LLVFRNADLD LDEMELHPYN SRGLFEKKLM QDNKQLYQIW 120
121 IVMVWLKENT YVMERPKNVP TSKWLNSITS GGLKSCDLDF PLRENTNVLD VKDKEEDHIF 180
181 FKYIYELILA GAIDEALEEA KLSDNISICM ILCGIQEYLN PVIDTQIANE FNTQQGIKKH 240
241 SLWRRTVYSL SQQAGLDPYE RAIYSYLSGA IPNQEVLQYS DWESDLHIHL NQILQTEIEN 300
301 YLLENNQVGT DELILPLPSH ALTVQEVLNR VASRHPSESE HPIRVLMASV ILDSLPSVIH 360
361 SSVEMLLDVV KGTEASNDII DKPYLLRIVT HLAICLDIIN PGSVEEVDKS KLITTYISLL 420
421 KLQGLYENIP IYATFLNESD CLEACSFILS SLEDPQVRKK QIETINFLRL PASNILRRTT 480
481 QRVFDETEQE YSPSNEISIS FDVNNIDMHL IYGVEWLIEG KLYVDAVHSI IALSRRFLLN 540
541 GRVKALEQFM ERNNIGEICK NYELEKIADN ISKDENEDQF LEEITQYEHL IKGIREYEEW 600
601 QKSVSLLSSE SNIPTLIEKL QGFSKDTFEL IKTFLVDLTS SNFADSADYE ILYEIRALYT 660
661 PFLLMELHKK LVEAAKLLKI PKFISEALAF TSLVANENDK IYLLFQSSGK LKEYLDLVAR 720
721 TATLSN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
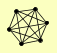
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
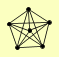
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
| View Details | Krogan NJ, et al. (2006) |
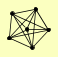
|
MEX67 NUP120 NUP145 NUP84 NUP85 SEH1 |
| View Details | Gavin AC, et al. (2006) |
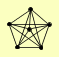
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
| View Details | Gavin AC, et al. (2002) |
|
CKA1 CKA2 CKB1 DBP2 MRPS28 NUP120 NUP145 NUP84 NUP85 RPP2B SEC13 SEC16 SEC23 SEC31 SEH1 STM1 |
| View Details | Ho Y, et al. (2002) |
|
ADE13 CBP3 HEM15 NUP120 NUP145 NUP84 NUP85 PHO86 SEC13 SEH1 YMR209C |
| View Details | Qiu et al. (2008) |
|
ASM4 GLE2 KAP120 MEX67 NIC96 NSP1 NUP1 NUP100 NUP116 NUP120 NUP133 NUP145 NUP157 NUP159 NUP188 NUP42 NUP49 NUP57 NUP82 NUP84 NUP85 POM152 SEC13 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
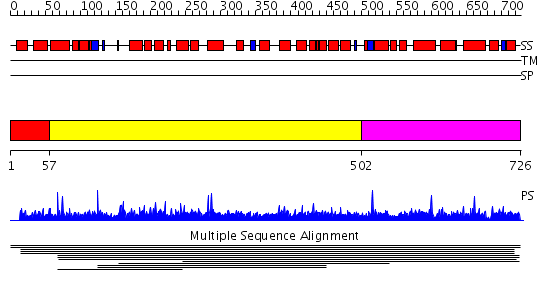
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..56] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [57..501] | 2.010903 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [502..726] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)