













| General Information: |
|
| Name(s) found: |
NUP145 /
YGL092W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1317 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[IDA |
| Biological Process: |
tRNA splicing, via endonucleolytic cleavage and ligation
[IMP protein export from nucleus [TAS ribosomal protein import into nucleus [TAS mRNA export from nucleus [IMP rRNA export from nucleus [TAS nuclear pore organization [IMP snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS protein import into nucleus [IMP chromosome localization [IMP tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS |
| Molecular Function: |
RNA binding
[IDA structural molecule activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFNKSVNSGF TFGNQNTSTP TSTPAQPSSS LQFPQKSTGL FGNVNVNANT STPSPSGGLF 60
61 NANSNANSIS QQPANNSLFG NKPAQPSGGL FGATNNTTSK SAGSLFGNNN ATANSTGSTG 120
121 LFSGSNNIAS STQNGGLFGN SNNNNITSTT QNGGLFGKPT TTPAGAGGLF GNSSSTNSTT 180
181 GLFGSNNTQS STGIFGQKPG ASTTGGLFGN NGASFPRSGE TTGTMSTNPY GINISNVPMA 240
241 VADMPRSITS SLSDVNGKSD AEPKPIENRR TYSFSSSVSG NAPLPLASQS SLVSRLSTRL 300
301 KATQKSTSPN EIFSPSYSKP WLNGAGSAPL VDDFFSSKMT SLAPNENSIF PQNGFNFLSS 360
361 QRADLTELRK LKIDSNRSAA KKLKLLSGTP AITKKHMQDE QDSSENEPIA NADSVTNIDR 420
421 KENRDNNLDN TYLNGKEQSN NLNKQDGENT LQHEKSSSFG YWCSPSPEQL ERLSLKQLAA 480
481 VSNFVIGRRG YGCITFQHDV DLTAFTKSFR EELFGKIVIF RSSKTVEVYP DEATKPMIGH 540
541 GLNVPAIITL ENVYPVDKKT KKPMKDTTKF AEFQVFDRKL RSMREMNYIS YNPFGGTWTF 600
601 KVNHFSIWGL VNEEDAEIDE DDLSKQEDGG EQPLRKVRTL AQSKPSDKEV ILKTDGTFGT 660
661 LSGKDDSIVE EKAYEPDLSD ADFEGIEASP KLDVSKDWVE QLILAGSSLR SVFATSKEFD 720
721 GPCQNEIDLL FSECNDEIDN AKLIMKERRF TASYTFAKFS TGSMLLTKDI VGKSGVSIKR 780
781 LPTELQRKFL FDDVYLDKEI EKVTIEARKS NPYPQISESS LLFKDALDYM EKTSSDYNLW 840
841 KLSSILFDPV SYPYKTDNDQ VKMALLKKER HCRLTSWIVS QIGPEIEEKI RNSSNEIEQI 900
901 FLYLLLNDVV RASKLAIESK NGHLSVLISY LGSNDPRIRD LAELQLQKWS TGGCSIDKNI 960
961 SKIYKLLSGS PFEGLFSLKE LESEFSWLCL LNLTLCYGQI DEYSLESLVQ SHLDKFSLPY1020
1021 DDPIGVIFQL YAANENTEKL YKEVRQRTNA LDVQFCWYLI QTLRFNGTRV FSKETSDEAT1080
1081 FAFAAQLEFA QLHGHSLFVS CFLNDDKAAE DTIKRLVMRE ITLLRASTND HILNRLKIPS1140
1141 QLIFNAQALK DRYEGNYLSE VQNLLLGSSY DLAEMAIVTS LGPRLLLSNN PVQNNELKTL1200
1201 REILNEFPDS ERDKWSVSIN VFEVYLKLVL DNVETQETID SLISGMKIFY DQYKHCREVA1260
1261 ACCNVMSQEI VSKILEKNNP SIGDSKAKLL ELPLGQPEKA YLRGEFAQDL MKCTYKI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
| View Details | Krogan NJ, et al. (2006) |
|
MEX67 NUP120 NUP145 NUP84 NUP85 SEH1 |
| View Details | Gavin AC, et al. (2006) |
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
| View Details | Gavin AC, et al. (2002) |

|
CKA1 CKA2 CKB1 DBP2 MRPS28 NUP120 NUP145 NUP84 NUP85 RPP2B SEC13 SEC16 SEC23 SEC31 SEH1 STM1 |
| View Details | Ho Y, et al. (2002) |
|
ADE13 NUP145 NUP84 NUP85 SEC13 SEH1 |
| View Details | Qiu et al. (2008) |
|
ASM4 GLE2 KAP123 MEX67 NDC1 NIC96 NSP1 NUP1 NUP100 NUP116 NUP120 NUP133 NUP145 NUP157 NUP159 NUP170 NUP188 NUP192 NUP2 NUP42 NUP49 NUP53 NUP57 NUP82 NUP84 NUP85 POM152 SEC13 SEH1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
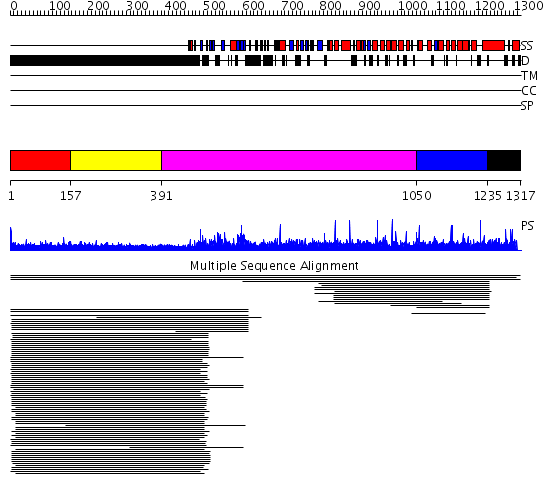
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | 5.69897 | Fibrinogen | |
| 2 | View Details | [157..390] | 19.626995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [391..1049] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1050..1234] | 2.012935 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1235..1317] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)