













| General Information: |
|
| Name(s) found: |
CDC15 /
YAR019C
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 974 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud neck
[TAS spindle pole body [IDA |
| Biological Process: |
regulation of exit from mitosis
[IMP cytokinesis [IMP protein amino acid phosphorylation [IDA |
| Molecular Function: |
protein kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSMADTDRV NLTPIQRASE KSVQYHLKQV IGRGSYGVVY KAINKHTDQV VAIKEVVYEN 60
61 DEELNDIMAE ISLLKNLNHN NIVKYHGFIR KSYELYILLE YCANGSLRRL ISRSSTGLSE 120
121 NESKTYVTQT LLGLKYLHGE GVIHRDIKAA NILLSADNTV KLADFGVSTI VNSSALTLAG 180
181 TLNWMAPEIL GNRGASTLSD IWSLGATVVE MLTKNPPYHN LTDANIYYAV ENDTYYPPSS 240
241 FSEPLKDFLS KCFVKNMYKR PTADQLLKHV WINSTENVKV DKLNKFKEDF TDADYHWDAD 300
301 FQEEKLNISP SKFSLRAAPA PWAENNQELD LMPPTESQLL SQLKSSSKPL TDLHVLFSVC 360
361 SLENIADTII ECLSRTTVDK RLITAFGSIF VYDTQHNHSR LRLKFIAMGG IPLIIKFEHL 420
421 AKEFVIDYPQ TLIECGIMYP PNFASLKTPK YILELVYRFY DLTSTAFWCR WCFKHLDISL 480
481 LLNNIHERRA QSILLKLSSY APWSFEKILP SLIDSKLKKK ILISPQITYV VFKSINYMIT 540
541 TNDDKIHKSA IPSSSSLPLS SSPTRNSPVN SVQSPSRSPV HSLMATRPSS PMRHKSISNF 600
601 PHLTISSKSR LLIELPEGFF TWLTSFFVDM AQIKDLSVLK YFTKLCYLTV HINSTFLNDL 660
661 LDNDAFFAFI RNIDTIIPFI DDAKTAAFIW KQITAICVEM SLDMDQMSAS LFSTAMNFIR 720
721 KKNNTSISGL EIILNCLHFT LRNVNDDVAP TVGSSESHSV FLIKVNNDAA IELPIDQLVD 780
781 LFYALNDDDV NLSKLISIFT KICSLPGFEN LTINIIFHPN FYEKIVSFFD TYFNSLLIQI 840
841 DLLKFIKLIF SKSLLKLYDY TGQPDPIKQT EPNRRNKATV FKLRAILVQI TEFLNNNWNN 900
901 GCPKRNSNQV GGDSVLICQL CEDIRSLSKK GSLQKVSSVT AAIGSSPTKD ERSNLRSSKD 960
961 KSDGFSVPIT TFQT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CDC15 DUG2 TEM1 |
| View Details | Krogan NJ, et al. (2006) |
|
BUD4 CDC13 CDC15 GIR2 PRS5 RBG2 YIL152W |
| View Details | Ho Y, et al. (2002) |

|
ATG4 BFA1 CAR2 CDC15 CDC33 CLU1 COF1 COR1 CPA2 CPR1 CPR3 CYS4 DBP8 DUG2 DUT1 FAA4 GCD11 GUS1 HEF3 KGD1 MCX1 NMD3 NUP53 OYE2 PFK1 PGM2 PST2 RNR1 RNR2 RPN1 SAR1 SEC26 SEC53 SSD1 TEF4 TEM1 TFP1 THI22 TIF1, TIF2 TMA29 UFD4 YDL086W YGR066C YHR033W YNK1 YTM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
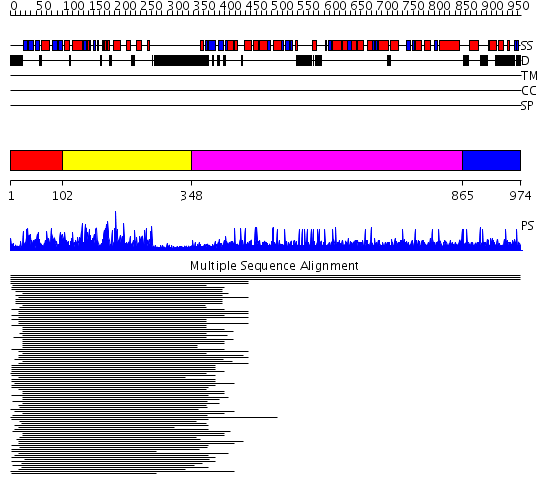
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 447.218487 | pak1 | |
| 2 | View Details | [102..347] | 447.218487 | pak1 | |
| 3 | View Details | [348..864] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [865..974] | 1.002947 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [758..871] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [872..974] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)