













| General Information: |
|
| Name(s) found: |
ACE2 /
YLR131C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 770 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
regulation of transcription involved in G1 phase of mitotic cell cycle
[IDA |
| Molecular Function: |
transcription activator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNVVDPWYI NPSGFAKDTQ DEEYVQHHDN VNPTIPPPDN YILNNENDDG LDNLLGMDYY 60
61 NIDDLLTQEL RDLDIPLVPS PKTGDGSSDK KNIDRTWNLG DENNKVSHYS KKSMSSHKRG 120
121 LSGTAIFGFL GHNKTLSISS LQQSILNMSK DPQPMELINE LGNHNTVKNN NDDFDHIREN 180
181 DGENSYLSQV LLKQQEELRI ALEKQKEVNE KLEKQLRDNQ IQQEKLRKVL EEQEEVAQKL 240
241 VSGATNSNSK PGSPVILKTP AMQNGRMKDN AIIVTTNSAN GGYQFPPPTL ISPRMSNTSI 300
301 NGSPSRKYHR QRYPNKSPES NGLNLFSSNS GYLRDSELLS FSPQNYNLNL DGLTYNDHNN 360
361 TSDKNNNDKK NSTGDNIFRL FEKTSPGGLS ISPRINGNSL RSPFLVGTDK SRDDRYAAGT 420
421 FTPRTQLSPI HKKRESVVST VSTISQLQDD TEPIHMRNTQ NPTLRNANAL ASSSVLPPIP 480
481 GSSNNTPIKN SLPQKHVFQH TPVKAPPKNG SNLAPLLNAP DLTDHQLEIK TPIRNNSHCE 540
541 VESYPQVPPV THDIHKSPTL HSTSPLPDEI IPRTTPMKIT KKPTTLPPGT IDQYVKELPD 600
601 KLFECLYPNC NKVFKRRYNI RSHIQTHLQD RPYSCDFPGC TKAFVRNHDL IRHKISHNAK 660
661 KYICPCGKRF NREDALMVHR SRMICTGGKK LEHSINKKLT SPKKSLLDSP HDTSPVKETI 720
721 ARDKDGSVLM KMEEQLRDDM RKHGLLDPPP STAAHEQNSN RTLSNETDAL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
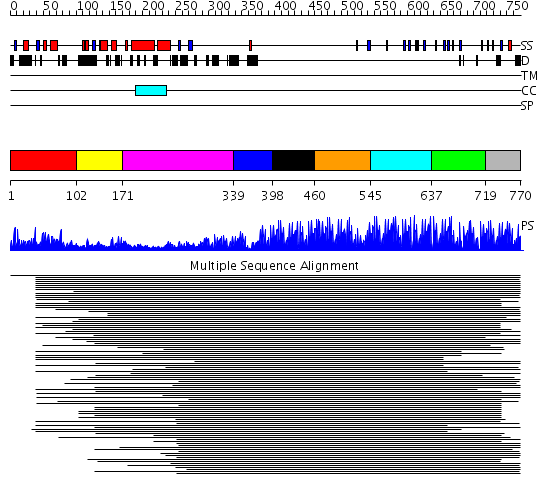
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 16.125996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [102..170] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [171..338] | 15.823996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [339..397] | 109.190949 | No description for 1lu6A_ was found. | |
| 5 | View Details | [398..459] | 109.190949 | No description for 1lu6A_ was found. | |
| 6 | View Details | [460..544] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [545..636] | 113.69897 | Ying-yang 1 (yy1, zinc finger domain) | |
| 8 | View Details | [637..718] | 1.970991 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [719..770] | 41.502279 | ADR1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)