













| General Information: |
|
| Name(s) found: |
p53-PB /
FBpp0083753
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 495 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC][IEA]
|
| Biological Process: |
determination of adult lifespan
[IMP]
response to radiation [IMP][TAS] DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis [IMP][TAS] regulation of transcription, DNA-dependent [IEA] positive regulation of cell cycle [IGI] DNA damage response, signal transduction resulting in induction of apoptosis [TAS] response to UV [IMP] response to DNA damage stimulus [IDA][IMP] regulation of transcription [IEA] tissue regeneration [IMP] regulation of transcription from RNA polymerase II promoter [TAS] cell death [IMP] positive regulation of gene-specific transcription [IMP] apoptosis [IMP][TAS] DNA damage response, signal transduction by p53 class mediator [TAS] activation of caspase activity [IMP] response to ionizing radiation [IMP] |
| Molecular Function: |
DNA binding
[NAS]
transcription factor activity [IEA][NAS] protein binding [IPI] RNA polymerase II transcription factor activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLHKSASFS LTFNQNTSIV SRSNSRTIFE AFKEFLDFWD IGNEVSAESA VRVSSNGAFN 60
61 LPQSFGNESN EYAHLATPVD PAYGGNNTNN MMQFTNNLEI LANNNSDGNN KINACNKFVC 120
121 HKGTDSEDDS TEVDIKEDIP KTVEVSGSEL TTEPMAFLQG LNSGNLMQFS QQSVLREMML 180
181 QDIQIQANTL PKLENHNIGG YCFSMVLDEP PKSLWMYSIP LNKLYIRMNK AFNVDVQFKS 240
241 KMPIQPLNLR VFLCFSNDVS APVVRCQNHL SVEPLTANNA KMRESLLRSE NPNSVYCGNA 300
301 QGKGISERFS VVVPLNMSRS VTRSGLTRQT LAFKFVCQNS CIGRKETSLV FCLEKACGDI 360
361 VGQHVIHVKI CTCPKRDRIQ DERQLNSKKR KSVPEAAEED EPSKVRRCIA IKTEDTESND 420
421 SRDCDDSAAE WNVSRTPDGD YRLAITCPNK EWLLQSIEGM IKEAAAEVLR NPNQENLRRH 480
481 ANKLLSLKKR AYELP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
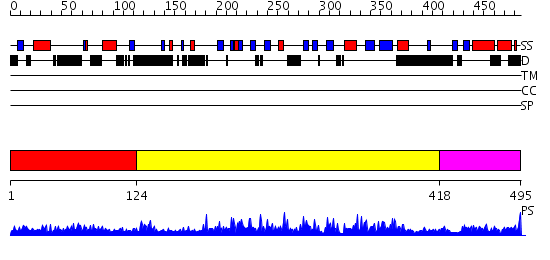
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | 1.034951 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [124..417] | 88.0 | Crystal structure of the human p53 core domain mutant M133L/V203A/N239Y/N268D at 1.9 A resolution. | |
| 3 | View Details | [418..495] | 5.064995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)