













| General Information: |
|
| Name(s) found: |
spn-E-PA /
FBpp0082637
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1434 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA][NAS] |
| Biological Process: |
intracellular mRNA localization
[IMP][TAS]
germarium-derived oocyte fate determination [IGI] polarity specification of dorsal/ventral axis [IMP] dorsal appendage formation [IMP] oocyte differentiation [NAS] chromatin silencing [IMP] pole plasm assembly [NAS] RNA localization [TAS] polarity specification of anterior/posterior axis [IMP] regulation of pole plasm oskar mRNA localization [IMP] oocyte maturation [IMP] oogenesis [IGI][TAS] karyosome formation [IMP] pole plasm oskar mRNA localization [IMP] RNA interference [TAS] oocyte localization during germarium-derived egg chamber formation [IMP] targeting of mRNA for destruction involved in RNA interference [IMP] |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] zinc ion binding [IEA] RNA helicase activity [TAS] RNA-dependent ATPase activity [TAS] helicase activity [ISS] ATP-dependent RNA helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQEVMDFFD FSKELKRVAA APQGYISSDP RLMATKFKSS EVPNRELIGT DYVSKIVAKE 60
61 KCLLNGTLLN EQPQGKRIRT LDDLDTDDEG EETEIRRDDE YYKKFRFNLN RDKNLSIYAK 120
121 REEILAAINA HPVVIIKGET GCGKTTQVPQ YILDEAYKSG KYCNIVVTQP RRIAAISIAN 180
181 RVCQEREWQQ NTVCSFQVGL HRPNSLEDTR LLYCTTGVLL NNLINNKTLT HYTHIVLDEV 240
241 HERDQNMDFL LIVVRRLLAT NSRHVKIILM SATIDAKELS DYFTTTNSIP PVITTNHRRK 300
301 HSIEKFYRDQ LGSIIWNEED VGHQQVPEIN KHGYRAAVKI IVIIDNMERK AAIQSRQSYD 360
361 EALRYGAVLI FLPGIYEIDT MAENLTCMLE NDPNIKVSIV RCFSLMTPEN QRDVFNPPPP 420
421 GFRKIILTTN IAESSITVPD VSYVIDFCLA KVKVTDTASS FSSLRLTWAS KANCRQRAGR 480
481 VGRLRSGRVY RMVNKHFYQR EMPEFGIPEM LRLPLQNSVL KAKVLNMGSP VEILALALSP 540
541 PNLSDIHNTI LLLKEVGALY LTVDGIYDPL DGDLTYWGTI MSRLPLDTRQ SRLIILGYIF 600
601 NMLEEAIIIA AGLSTPGLFA HEGGRSQLGD SFWMHYIFSD GSGSDLVAIW RVYLTYLNIV 660
661 ENGHDQESAI RWAKRFHVSL RSLKEIHLLV QELRVRCTHL GLIPFPVNPN QMMDDREKAI 720
721 MLKVIIAGAF YPNYFTRSKE SCADTDRNIY QTISGHDPCR TVYFTNFKPA YMGELYTRRI 780
781 KELFQEVRIP PENMDVTFQE GSQKVFVTFK QDDWIEGSSK YVPVSGRVQS EVYKAVMMRQ 840
841 NRVERPIHIM NPSAFMSYVQ QRGIGDVIEG RWIPPTKPLN VELLALPSVF DKTISGSITC 900
901 IVNCGKFFFQ PQSFEECIRN MSEIFNAPQQ LRNYVTNASA IAKGMMVLAK RDSYFQRATV 960
961 IRPENQSNRQ PMFYVRFIDY GNCTLLPMQL MRLMPRELTE QYGDLPPRVF ECRLAMVQPS1020
1021 SVVSGNNRWS TAANDMLKTV AQCGLIDIEV YSLFNNVAAV LIHMRDGIIN DKLVELMLCR1080
1081 RSDEDYMSRK DHDFRLRRQE SARNLSTAQR QQINEEYLRS CQLPQDHDLP PPPLEKCKTV1140
1141 VMLKGPNSPL ECTMRSITRV GLSKRVNIDH LSVNALLLDA DPQDHHDHLI VAHEIAESRN1200
1201 GQTLTARGTT LMPNVQGFGA LMVMLFSPTM QLKCNKEGTS YVSVLGGLGC DPDTNEPYFA1260
1261 EHDVLINLDV NILEDDVILI NQIRYYIDSV FFNFKEENNP AVSVNERVSI YTQLRSLINR1320
1321 LLCKDRRYIE RNMSNADFEW ETNPELPLPN EPFGKRAIFP MHSLTELQEE DTGRLVQLRE1380
1381 NCSMLHKWRN FEGTLPHMTC KLCNQLLESV PQLRLHLLTI LHRDREKQID YCNQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
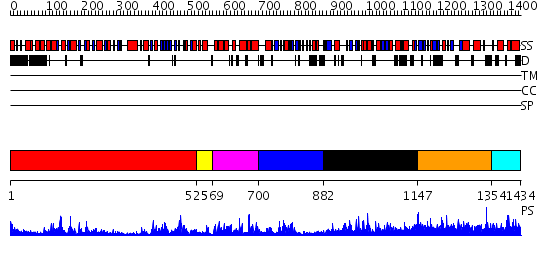
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..524] | 62.39794 | No description for 2hyiC was found. | |
| 2 | View Details | [525..568] | 7.0 | Nucleotide excision repair enzyme UvrB | |
| 3 | View Details | [569..699] | 1.13 | No description for 2v8oA was found. | |
| 4 | View Details | [700..881] | 1.54999 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [882..1146] | 28.39794 | No description for 2hqeA was found. | |
| 6 | View Details | [1147..1353] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1354..1434] | 1.29 | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)