













| General Information: |
|
| Name(s) found: |
gi|7303012
[NCBI NR]
gi|24653990 [NCBI NR] gi|24653988 [NCBI NR] gi|21645324 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1086 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA]
cytoplasm [ISS] |
| Biological Process: |
cellular carbohydrate metabolic process
[IEA]
acetyl-CoA biosynthetic process [ISS] citrate metabolic process [ISS] |
| Molecular Function: |
ATP binding
[IEA]
ATP citrate synthase activity [ISS][NAS] succinate-CoA ligase (ADP-forming) activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAKAITEAS GKDILNRHLN THGAGAATCR FSTVNSTTDW SKLAVDHPWL LTTPLVCKPD 60
61 QLIKRRGKLG LIGVKKNFEQ VKQWIGERLN KDQKIGNAVG KLRNFIIEPF VPHTDAEEMY 120
121 VCIYSHRAAD TILFYHQGGV DIGDVDAKAV KLDVPVNSSL SLADVKSKLL KEVKDAGTKE 180
181 RIAKFVSALY TTYVDLYFTY LEINPLVVTA DNLYILDLAA KLDSTADFIC RPKWGEIDYP 240
241 PPFGRDAYPE EAYIADLDAK SGASLKLTIL NRNGRIWTMV AGGGASVIYS DTICDLGGAS 300
301 ELANYGEYSG APSEQQTYEY AKTILNLMTS SPKHPDGKVL ITGGGIANFT NVAATFQGII 360
361 TALREFQPKL VEHNVSIFVR RAGPNYQEGL RKMRDFGSTL GIPLHVFGPE THMTAICGMA 420
421 LGKRPIPQTA SVEFSTANFL LPGGQQAQAD LKAASDASEA LGSGSADEAD SAGISGAQRN 480
481 GSSLNRKFFS NTTKAIVWGM QQRAVQRDEP SVVAMVYPFT GDHKQKYYWG HKEILIPVYK 540
541 KMSDAIHKHK EVDVMVNFAS MRSAYESTLE VLEFPQIRTV AIIAEGIPEN MTRKLIIEAD 600
601 KKGVAIIGPA TVGGVKPGCF KIGNTGGMLD NILHSKLYRP GSVAYVSRSG GMSNELNNII 660
661 SKATDGVIEG IAIGGDRYPG STFMDHILRY QADPETKLIV LLGEVGGTEE YDVCAALKDG 720
721 RITKPLVAWC IGTCASMFTS EVQFGHAGSC ANSDRETATA KNKGLRDAGA YVPDSFDTLG 780
781 ELIHHVYGEL VKTGRVVPKE EVPPPTVPMD YSWARELGLI RKPASFMTSI CDERGQELIY 840
841 AGMPISEVLS KDVGIGGVIS LLWFQRCLPS YVCKFFEMCL MVTADHGPAV SGAHNTIVCA 900
901 RAGKDLVSSV VSGLLTIGDR FGGALDGSAR QFSEAYDTNL HPMEFVNKMR KEGKLILGIG 960
961 HRVKSINNPD VRVKIIKEFV LENFPACPLL KYALEVEKIT TNKKPNLILN VDGVIATAFV1020
1021 DMLRNSGSFT SEEAQEYINV GAINSLFVLG RSIGFIGHYM DQKRLKQGLY RHPWDDISYV1080
1081 IPEQYN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
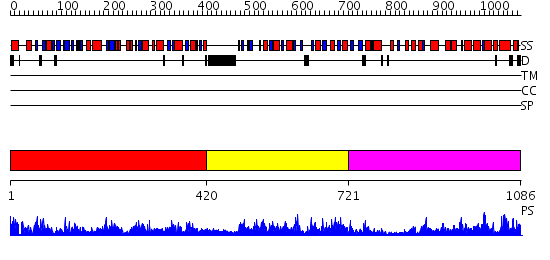
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..419] | 102.0 | Succinyl-CoA synthetase, beta-chain, C-terminal domain; Succinyl-CoA synthetase, beta-chain, N-terminal domain | |
| 2 | View Details | [420..720] | 93.69897 | Succinyl-CoA synthetase, alpha-chain, N-terminal (CoA-binding) domain; Succinyl-CoA synthetase, alpha-chain, C-terminal domain | |
| 3 | View Details | [721..1086] | 100.0 | Citrate synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)