













| General Information: |
|
| Name(s) found: |
PLC1 /
YPL268W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 869 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA condensed nuclear chromosome kinetochore [IDA |
| Biological Process: |
signal transduction
[IMP regulation of phosphate metabolic process [IMP pseudohyphal growth [IMP vacuole fusion, non-autophagic [IMP |
| Molecular Function: |
phosphoinositide phospholipase C activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTESAIDDQR FNLTKELQRH SCRDQGKITQ KDDALDFISY SSFQSSFNTD QKSANNGSTV 60
61 RRSIRSIFRR AAELPRVHMG PLTYSHGINE LVNKKLRKDC DLSTLCRVLQ RGIRMIRMTR 120
121 RRRKFYEFKL INNNGQIIWK DGSKYLELDS VKDIRIGDTA STYQEEVDPK RLRSDSKLWI 180
181 AIIYKVSNKL KALHVVALNE LDFNTFLSCI CGLVKLRREL MESILLPDNS QFARIHWQIT 240
241 VSEKEEDEKK DTLSFADVKK LCDKFHIYVS TGQLLEFFQL ADINHNGLLN YFEFEKFIKI 300
301 LKNRKEVNMI WSKFTKPPHS HLSFENFFQF LITEQHEQVD RQTAWSYFIK YREPTQLTMG 360
361 QDGFTKFLKE QPYLVEVKEE LYSKPLNHYF IASSHNTYLL GKQIAETPSV EGYIQVLQQG 420
421 CRCVEIDIWD GENGPVVCHG FLTSAIPLKT VIRVIKKYAF ITSPYPLIIS LEINCNKDNQ 480
481 KLASLIMREV LAEQLYFVGT RTDKLPSPRE LKHKILLKSK KTSEATRGLS VNEPFPSSFS 540
541 SSYESANEQE LRMKDDSTNS SSATNSSSMQ RIKRIGLKKH ADIINDVSNI SGIHGIKFRN 600
601 FSLPESKTIA HCFSLNERKV EYMIKDKHLK LSLDKHNRRY LMRVYPHVLR YKSSNFNPIP 660
661 FWKAGVQMVA TNWQTNDIGQ QLNLAMFQIL DHQPDGSFKS GYVLKPKKLL PVVTKAKMIP 720
721 LIYEHFENGS DPVTVKIRIL STQLLPRLND TSPSRNNTNS FVKVEFHTDD EPTMPISIDK 780
781 GTRISATEAS TKSSQGNGFN PIWDAEVSIT LKDTDLTFIK FMVISEETQI ASVCLKLNYL 840
841 RMGYRHIPLF NMEGEQYIFC TLFIHTQIL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BTT1 CAF130 EGD1 EGD2 PLC1 YJR011C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
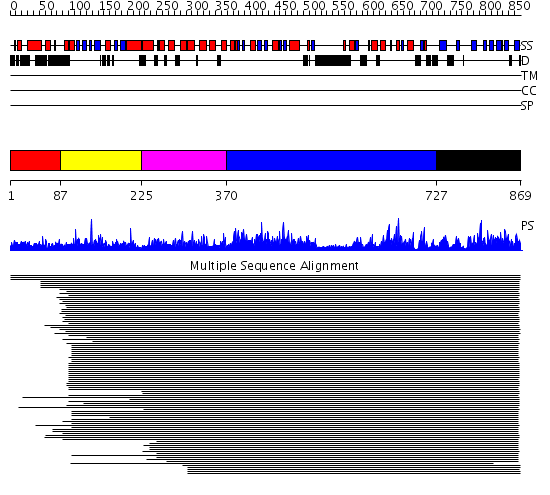
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..224] | 20.09691 | Phospholipase C delta-1 | |
| 3 | View Details | [225..369] | 1576.9897 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) | |
| 4 | View Details | [370..726] | 1576.9897 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) | |
| 5 | View Details | [727..869] | 1576.9897 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)