













| General Information: |
|
| Name(s) found: |
NCS2 /
YNL119W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 493 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
protein modification process
[IMP pseudohyphal growth [IMP ribosome biogenesis [RCA invasive growth in response to glucose limitation [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MECQRCPASA RNPATVESRK EKFCDECFIK FVSTKQRKQM MKDEYFRNLF KVIYPFEKEG 60
61 SVSKILLPLS HSDSGSLVML DIVHDLLLEQ TKQHNNRTGF TVDVLTVFTE ENVSVIKERM 120
121 ESLINEKMSQ LNKISNIFNV HFIDVNEFFN NASEVSTFII DNENFEIFSK SKSVDDSNIL 180
181 TLKEILGKYC LNNSSRSDLI SIIKTQLIKH FAYENGYNAI MWGHSMTKLS EVIISLVVKG 240
241 KGSQIATFLD SESFDTLNNK PCKYKNLYPM KDLLSVEIES FLQIRNLAQF LINVEETNVK 300
301 PNCLIARKSL PSLGQQKLVK NMTINEITNK YFQDIQNDYS NIISTVLRTA DKLTQPKSSM 360
361 AKPSQCQICQ SKIYTNPSNW LNRITVTSPY PVETTEEKYL FKQWQDSKLG QSHTHYVELL 420
421 NEIKQGASNS LDVEDGDVKL CYGCLILLNT SIKDKNLVWP KVDTMDITAN ATNKNKELSQ 480
481 ILDQFEINSD GEE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
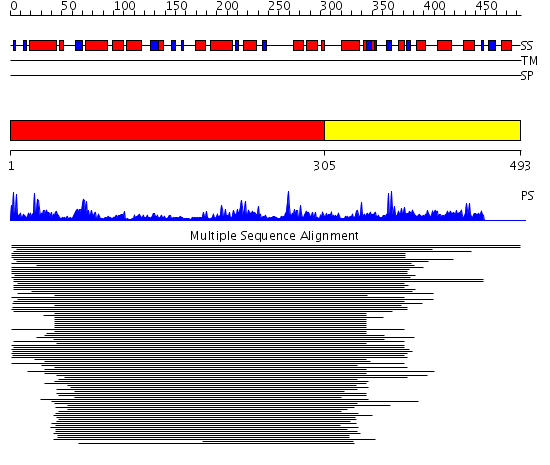
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..304] | 9.88 | Argininosuccinate synthetase, N-terminal domain; Argininosuccinate synthetase, C-terminal domain | |
| 2 | View Details | [305..493] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)