













| General Information: |
|
| Name(s) found: |
DSS1 /
YMR287C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 969 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA mitochondrion [IDA |
| Biological Process: |
RNA catabolic process
[IDA |
| Molecular Function: |
exoribonuclease II activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVRRKVHVL LIARSFHSYT PCFRVTTRGK RQRSKSKQQA KVELDHTREL DNDQATETVV 60
61 DRSVGPEKDI ESINKDFLQR TKGLEPDIEL KQLPQIKQEF NQRYKDRYVK PSEDWYVNSW 120
121 RSLTKPKIPL YKLINSDFQL ITKLKAPNPM EFQPVQLMES PLNVGDFVLL KMRPNELAMC 180
181 VSLPSSTMDP RYTFVTIDGT MCFATKNRVL LRIPHKLPAG IHSLIQPESH HKHLPIGTVK 240
241 NFSNQTNILP IVARQLITSR YPAQISKLAW KDLPITTKKL QLLHRSLQNY MGPWQIPFFT 300
301 LVGLVQKLDL NKALDDKNGI NYLTSLVNNY HTVNDIPINS PTFVSTYWAI MQQQESNLWG 360
361 EIHLNTALLS PISVTIIPLK SQHLYYAQVI EKLEANSYRE VNKFVKLVNE RKYRDISALY 420
421 PSVIQLLKDF AAGNFHNNGI IVALISKIFR KIERYKDCDI TRDICQDLIN EITPNSIPNP 480
481 LLLNMDLALP ASSKLVKWQQ KLYDLTNIEE LQWKKSGTDD DRYDFGDLRV FCIDSETAHE 540
541 IDDGVSVKNY GRDGLYTLYI HIADPTSMFP ESTNVDIEGI STDILNVALK RSFTTYLPDT 600
601 VVPMLPQSIC HLSDLGKQGQ RTKTISFSVD VKITSKCSGK SIEIMYDSFK IRKGIVSNFP 660
661 KATYEDVDRI LGTPNSEASP VKKDLESLSM ISKLLREQRI KNSNAVIFGE GFNKGLVMLN 720
721 ADSEGELTEV TFSDQEETLS TILVSEMMIL ANTLTGRYFA ENKIGGVFRC YKQLPLGEVA 780
781 QQQYDSMITS TKKGIFPKLK DIVKLSSLLN SSFYTGRPFR HEMIGAKQYL TVTSPLRRFP 840
841 DLINHLQIHR HLQKKPLCFN QTQIDSLIWP IQSRADILKR ASRNSSTYWT LNYLKKLTKL 900
901 EPERTFDVMV TSVPQNGFTG CVFPDLSFAR GTLKLHPSSM HYPMIGDIVK NCKISKIDCL 960
961 EGMLELEKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
DSS1 SUV3 |
| View Details | Krogan NJ, et al. (2006) |

|
CSE2 DSS1 GAL11 GPT2 IRC11 MED1 MED11 MED2 MED4 MED6 MED7 MED8 NUT1 NUT2 PGD1 RGR1 ROX3 SIN4 SOH1 SRB2 SRB4 SRB5 SRB6 SRB7 SSN3 SSN8 THI72 |
| View Details | Ho Y, et al. (2002) |
|
ADH2 APT1 ARO1 CKA1 COP1 DSS1 ECM10 MAM33 MTC5 OYE2 PRE1 PST2 PUP3 SLD2 SOD2 SRP1 STI1 TMA29 TPS1 TRL1 TSA2 YHR033W YKU80 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
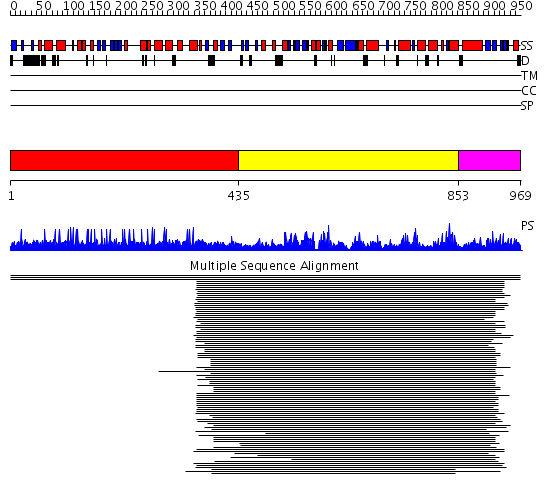
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..434] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [435..852] | 115.236572 | RNB-like protein No confident structure predictions are available. | |
| 3 | View Details | [853..969] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)