













| General Information: |
|
| Name(s) found: |
MRE11 /
YMR224C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 692 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA Mre11 complex [IPI |
| Biological Process: |
DNA repair
[IMP meiotic DNA double-strand break processing [TAS double-strand break repair via break-induced replication [IGI telomere maintenance [IMP double-strand break repair via nonhomologous end joining [IMP meiotic DNA double-strand break formation [TAS |
| Molecular Function: |
adenylate kinase activity
[IDA 3'-5' exonuclease activity [IDA protein binding [IPI telomeric DNA binding [IDA endonuclease activity [IDA endodeoxyribonuclease activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDYPDPDTIR ILITTDNHVG YNENDPITGD DSWKTFHEVM MLAKNNNVDM VVQSGDLFHV 60
61 NKPSKKSLYQ VLKTLRLCCM GDKPCELELL SDPSQVFHYD EFTNVNYEDP NFNISIPVFG 120
121 ISGNHDDASG DSLLCPMDIL HATGLINHFG KVIESDKIKV VPLLFQKGST KLALYGLAAV 180
181 RDERLFRTFK DGGVTFEVPT MREGEWFNLM CVHQNHTGHT NTAFLPEQFL PDFLDMVIWG 240
241 HEHECIPNLV HNPIKNFDVL QPGSSVATSL CEAEAQPKYV FILDIKYGEA PKMTPIPLET 300
301 IRTFKMKSIS LQDVPHLRPH DKDATSKYLI EQVEEMIRDA NEETKQKLAD DGEGDMVAEL 360
361 PKPLIRLRVD YSAPSNTQSP IDYQVENPRR FSNRFVGRVA NGNNVVQFYK KRSPVTRSKK 420
421 SGINGTSISD RDVEKLFSES GGELEVQTLV NDLLNKMQLS LLPEVGLNEA VKKFVDKDEK 480
481 TALKEFISHE ISNEVGILST NEEFLRTDDA EEMKALIKQV KRANSVRPTP PKENDETNFA 540
541 FNGNGLDSFR SSNREVRTGS PDITQSHVDN ESRITHISQA ESSKPTSKPK RVRTATKKKI 600
601 PAFSDSTVIS DAENELGDNN DAQDDVDIDE NDIIMVSTDE EDASYGLLNG RKTKTKTRPA 660
661 ASTKTASRRG KGRASRTPKT DILGSLLAKK RK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
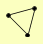
MRE11 XRS2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
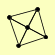
MRE11 RAD50 XRS2 |
| View Details | Krogan NJ, et al. (2006) |
|
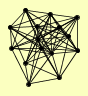
DMC1 MRE11 RAD50 XRS2 YBL044W |
| View Details | Ho Y, et al. (2002) |
|
AHP1 CDC16 DUN1 ERG20 GPH1 MAM33 MKT1 MRE11 RAD50 REX2 RPT3 SEC27 TFP1 VMA8 XRS2 |
| View Details | Qiu et al. (2008) |
|
MRE11 XRS2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
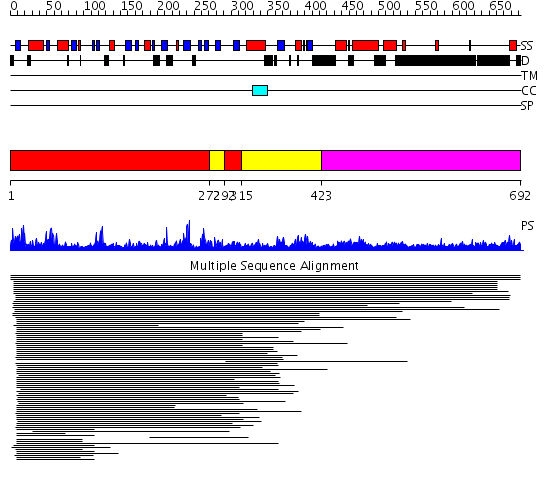
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..271] [292..314] |
100.10721 | Mre11 | |
| 2 | View Details | [272..291] [315..422] |
100.10721 | Mre11 | |
| 3 | View Details | [423..692] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)