













| General Information: |
|
| Name(s) found: |
ISA1 /
YLL027W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 250 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA |
| Biological Process: |
iron ion transport
[IGI telomere maintenance [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MINTGRSRNS VLLAHRFLST GGFWRGGTNG TMSRTINNVN PFKLKFIPKT VPAAADSVSP 60
61 DSQRPGKKPF KFIVSNQSKS SKASKSPKWS SYAFPSRETI KSHEEAIKKQ NKAIDEQIAA 120
121 AVSKNDCSCT EPPKKRKRKL RPRKALITLS PKAIKHLRAL LAQPEPKLIR VSARNRGCSG 180
181 LTYDLQYITE PGKFDEVVEQ DGVKIVIDSK ALFSIIGSEM DWIDDKLASK FVFKNPNSKG 240
241 TCGCGESFMV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ISA1 NFU1 |
| View Details | Ho Y, et al. (2002) |

|
CBF5 CBP2 CCM1 CDC33 CIC1 DBP7 ENP2 GBP2 HAS1 HMO1 HTB2 IMD1 IMD3 ISA1 KRE33 KRI1 MGM101 MSS116 NOC2 NOP12 NOP6 NPL3 PET127 POL5 PRP43 PUF6 PWP1 RLI1 RRP12 RRP5 TIF4631 TRA1 TSR1 URB1 URB2 UTP10 UTP20 YPP1 YRA1 YTM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
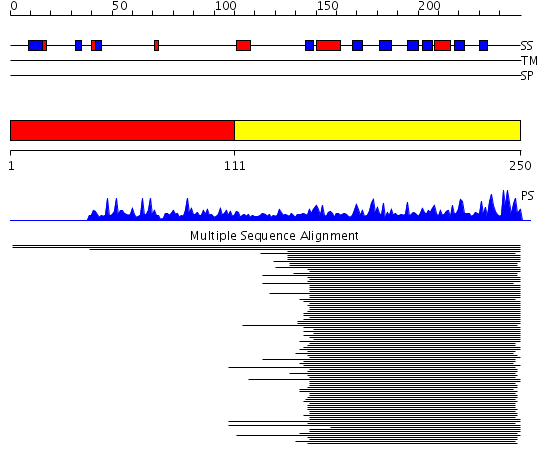
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [111..250] | 50.318759 | Iron-sulphur cluster biosynthesis No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)