













| General Information: |
|
| Name(s) found: |
GPM1 /
YKL152C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 247 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA cytosol [IDA |
| Biological Process: |
gluconeogenesis
[IMP glycolysis [IMP |
| Molecular Function: |
phosphoglycerate mutase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKLVLVRHG QSEWNEKNLF TGWVDVKLSA KGQQEAARAG ELLKEKKVYP DVLYTSKLSR 60
61 AIQTANIALE KADRLWIPVN RSWRLNERHY GDLQGKDKAE TLKKFGEEKF NTYRRSFDVP 120
121 PPPIDASSPF SQKGDERYKY VDPNVLPETE SLALVIDRLL PYWQDVIAKD LLSGKTVMIA 180
181 AHGNSLRGLV KHLEGISDAD IAKLNIPTGI PLVFELDENL KPSKPSYYLD PEAAAAGAAA 240
241 VANQGKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
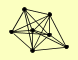
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ADH1 EFT2, EFT1 ENO2 FBA1 GPM1 HSC82 TPI1 |
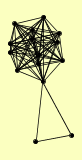
| View Details | Gavin AC, et al. (2002) |
|
ADH1 ENO2 FAR11 FAR3 FBA1 GPM1 HFM1 HSC82 PDC1 RAM1 RAM2 ROM2 SNT2 SSE2 TPI1 |
| View Details | Qiu et al. (2008) |

|
GPM1 PGM2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
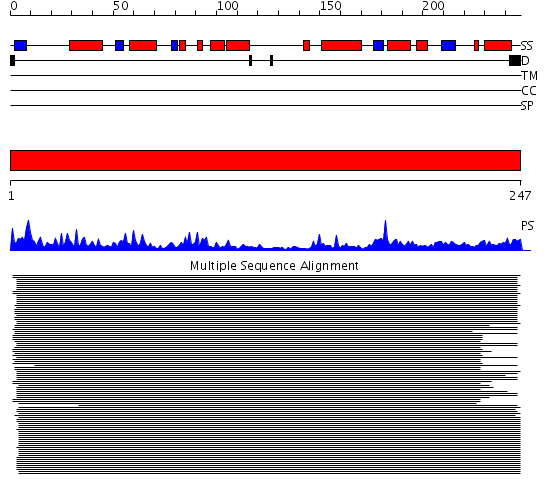
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..247] | 1387.09691 | Phosphoglycerate mutase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)