













| General Information: |
|
| Name(s) found: |
RAD2 /
YGR258C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1031 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleotide-excision repair factor 3 complex
[IDA |
| Biological Process: |
nucleotide-excision repair, DNA incision, 3'-to lesion
[TAS nucleotide-excision repair [IDA |
| Molecular Function: |
single-stranded DNA specific endodeoxyribonuclease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVHSFWDIA GPTARPVRLE SLEDKRMAVD ASIWIYQFLK AVRDQEGNAV KNSHITGFFR 60
61 RICKLLYFGI RPVFVFDGGV PVLKRETIRQ RKERRQGKRE SAKSTARKLL ALQLQNGSND 120
121 NVKNSTPSSG SSVQIFKPQD EWDLPDIPGF KYDKEDARVN SNKTFEKLMN SINGDGLEDI 180
181 DLDTINPASA EFEELPKATQ YLILSSLRLK SRLRMGYSKE QLETIFPNSM DFSRFQIDMV 240
241 KRRNFFTQKL INTTGFQDGG ASKLNEEVIN RISGQKSKEY KLTKTNNGWI LGLGANDGSD 300
301 AQKAIVIDDK DAGALVKQLD SNAEDGDVLR WDDLEDNSLK IVRHESSNAT TAPQKRSNRS 360
361 EDEGCDSDEC EWEEVELKPK NVKFVEDFSL KAARLPYMGQ SLNNAGSKSF LDKRHDQASP 420
421 SKTTPTMRIS RISVEDDDED YLKQIEEIEM MEAVQLSKME KKPEADDKSK IAKPVTSKGT 480
481 EARPPIVQYG LLGAQPDSKQ PYHVTNLNSK SESVIKRTSK TVLSEFRPPS QQEDKGAILT 540
541 EGEQNLNFIS HKIPQFDFNN ENSLLFQKNT ESNVSQEATK EKSPIPEMPS WFSSTASQQL 600
601 YNPYNTTNFV EDKNVRNEQE SGAETTNKGS SYELLTGLNA TEILERESEK ESSNDENKDD 660
661 DLEVLSEELF EDVPTKSQIS KEAEDNDSRK VESINKEHRK PLIFDYDFSE DEEDNIVENM 720
721 IKEQEEFDTF KNTTLSTSAE RNVAENAFVE DELFEQQMKD KRDSDEVTMD MIKEVQELLS 780
781 RFGIPYITAP MEAEAQCAEL LQLNLVDGII TDDSDVFLFG GTKIYKNMFH EKNYVEFYDA 840
841 ESILKLLGLD RKNMIELAQL LGSDYTNGLK GMGPVSSIEV IAEFGNLKNF KDWYNNGQFD 900
901 KRKQETENKF EKDLRKKLVN NEIILDDDFP SVMVYDAYMR PEVDHDTTPF VWGVPDLDML 960
961 RSFMKTQLGW PHEKSDEILI PLIRDVNKRK KKGKQKRINE FFPREYISGD KKLNTSKRIS1020
1021 TATGKLKKRK M |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
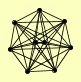
|
CCL1 KIN28 RAD2 RAD3 SSL1 SSL2 TFB1 TFB2 TFB3 |
| View Details | Ho Y, et al. (2002) |

|
PEX15 RAD2 |
| View Details | Qiu et al. (2008) |
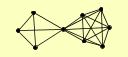
|
CCL1 RAD1 RAD16 RAD2 RAD3 RAD7 SSL1 SSL2 TFB3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
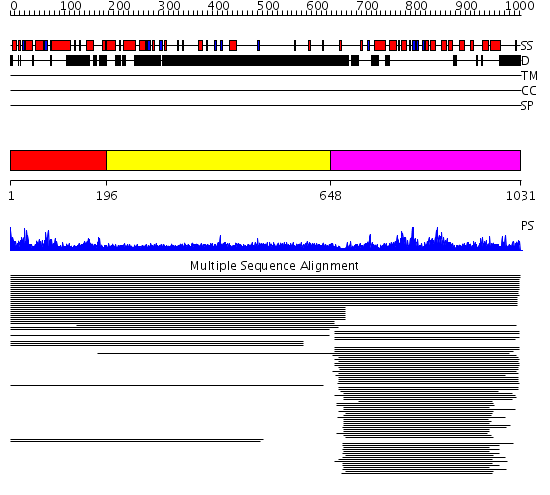
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..195] | 12.55 | 5' to 3' exonuclease domain of DNA polymerase Taq; Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) | |
| 2 | View Details | [196..647] | 32.249997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [648..1031] | 166.9897 | Flap endonuclease-1 (Fen-1 nuclease) | |
| 4 | View Details | [671..1031] | 72.522879 | Crystal structure of the human FEN1-PCNA complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)