













| General Information: |
|
| Name(s) found: |
MET10 /
YFR030W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1035 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[TAS sulfite reductase complex (NADPH) [IDA |
| Biological Process: |
response to drug
[IMP sulfate assimilation [TAS |
| Molecular Function: |
sulfite reductase (NADPH) activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPVEFATNPF GEAKNATSLP KYGTPVTAIS SVLFNNVDSI FAYKSFSQPD LLHQDLKKWS 60
61 EKRGNESRGK PFFQELDIRS GAGLAPLGFS HGLKNTTAIV APGFSLPYFI NSLKTVSHDG 120
121 KFLLNVGALN YDNATGSVTN DYVTALDAAS KLKYGVVTPI SANEVQSVAL LALAIATFSN 180
181 NSGAINLFDG LNYSKTVLPL VESVPEASIL AKLSKVIAPD AAFDDVLDKF NELTGLRLHN 240
241 FQYFGAQDAE TVFITYGSLE SELFNSAISG NNSKIGLINV RVPLPFNVAK FVTHVPSTTK 300
301 QIVVIGQTLD GSSPSFLRSQ VSAALFYHGR TSISVSEYIY QPDFIWSPKA VKSIVSSFIP 360
361 EFTYNADSSF GEGFIYWASD KSINIDVASK LVKALSLEDG KFVSLRTKFD NLANAGTFQA 420
421 QFVTSKEQIP VSNIDSTKLS VVEDVSLLKH LDVAATVAEQ GSIALVSQKA VKDLDLNSVE 480
481 SYVKNLGIPE SFLISIAKKN IKLFIIDGET TNDESKLSLF IQAVFWKLAF GLDVAECTNR 540
541 IWKSIDSGAD ISAASISEFL TGAFKNFLSE VPLALYTKFS EINIEKKEDE EEPAALPIFV 600
601 NETSFLPNNS TIEEIPLPET SEISDIAKKL SFKEAYEVEN KLRPDLPVKN FVVKVKENRR 660
661 VTPADYDRYI FHIEFDISGT GMTYDIGEAL GIHARNNESL VKEFLTFYGL NESDVVLVPN 720
721 KDNHHLLETR TVLQAFVENL DIFGKPPKRF YESLIPYASN EEEKKKLEDL VTPAGAVDLK 780
781 RFQDVEYYTY ADIFELFPSV RPSLEELVTI IEPLKRREYS IASSQKVHPN EVHLLIVVVD 840
841 WVDNKGRKRY GQASKYISDL AVGSELVVSV KPSVMKLPPS PKQPVIMSGL GTGLAPFKAI 900
901 VEEKLWQKQQ GYEIGEVFLY LGSRHKREEY LYGELWEAYK DAGIITHIGA AFSRDQPQKI 960
961 YIQDRIKENL DELKTAMIDN KGSFYLCGPT WPVPDITQAL QDILAKDAEE RGIKVDLDAA1020
1021 IEELKEASRY ILEVY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ALG5 BRE5 CSN12 CUP2 DBP7 LUG1 MET10 MSC7 MSE1 NOP53 NPY1 PBP4 RPF2 RRS1 SAS5 UME6 YGR126W YHR127W YPR045C YRA1 |
| View Details | Gavin AC, et al. (2006) |
|
MET10 RPA135 |
| View Details | Ho Y, et al. (2002) |
|
ATG12 CDC28 CLU1 CRM1 FET3 GSY1 KAP122 KGD1 MET10 MET18 PFK1 RPN1 TCP1 |
| View Details | Qiu et al. (2008) |

|
MET10 MET5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
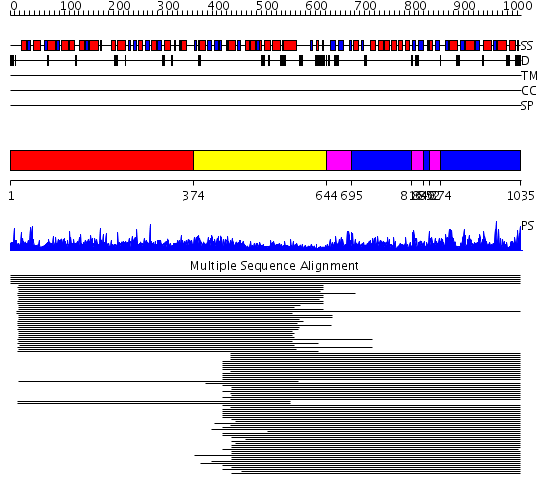
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..373] | 231.9897 | Pyruvate-ferredoxin oxidoreductase, PFOR, domain I; Pyruvate-ferredoxin oxidoreductase, PFOR, domains VI; Pyruvate-ferredoxin oxidoreductase, PFOR, domain II; Pyruvate-ferredoxin oxidoreductase, PFOR, domain III; Pyruvate-ferredoxin oxidoreductase, PFOR, domain V | |
| 2 | View Details | [374..643] | 231.9897 | Pyruvate-ferredoxin oxidoreductase, PFOR, domain I; Pyruvate-ferredoxin oxidoreductase, PFOR, domains VI; Pyruvate-ferredoxin oxidoreductase, PFOR, domain II; Pyruvate-ferredoxin oxidoreductase, PFOR, domain III; Pyruvate-ferredoxin oxidoreductase, PFOR, domain V | |
| 3 | View Details | [644..694] [816..839] [852..873] |
618.54902 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain | |
| 4 | View Details | [695..815] [840..851] [874..1035] |
618.54902 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)