













| General Information: |
|
| Name(s) found: |
MTC5 /
YDR128W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1148 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole membrane
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCSSINEGPY NSPTFGKSLS LKVDGGFNAV SINPSGRDIV LASRQGLYII DLDDPFTPPR 60
61 WLHHITPWQV ADVQWSPHPA KPYWIVSTSN QKAIIWNLAK SSSNAIEFVL HGHSRAITDI 120
121 NFNPQHPDVL ATCSVDTYVH AWDMRSPHRP FYSTSSWRSA ASQVKWNYKD PNVLASSHGN 180
181 DIFVWDLRKG STPLCSLKGH VSSVNSIDFN RFKYSEIMSS SNDGTVKFWD YSKSTTESKR 240
241 TVTTNFPIWR GRYLPFGEGY CIMPMVGGNN AVYLINLCDD DDSEQNKKTK LQPIYAFKGH 300
301 SDRVIDFLWR SRHTCDGDYD DREFQLVTWS KDCDLKLWPI SDSIYGKVNF DRGKRLEEKL 360
361 PDYDYCSYNK EPENRENVQK NEFRRLRENF VTTSGLKKNK TNHITWLSGI RMNSATSQED 420
421 LFNETKIQNL GEEVSAIGHK FPKVVFEKIS VSTRELCLTL NGPWSEENPD DYIFLRISIN 480
481 FPLNYPNKGD PPKFTIEENS NLTMSKRQEI LSNLATIGQK YTDSNLYCLE PCIRFVLGEK 540
541 VSLEDIEEGQ EPLLNFDIAD HIDFEELSSL DSSYSDSQNP ENLSSQSDIE SYKEALVFPD 600
601 TSNQGLDFGR NLALDTTPVP NGCGSCWTAT GELFCFFANE KKPEKKQNAI IKLSQKEAGV 660
661 EKHPFKIEPQ VLYDKEVDSS VITAADELKA RPKRYVDTLG LGGGTNGDSR TYFDDETSSD 720
721 DSFDSVADDW DDILRNDIIV RTKIPILRGN FKAFSSVHSE SGKTVESTKK NKNLVISKNF 780
781 SSLLSDRKEL ALEYLFMDAT PEGFARNNAL VAEKFDLDEI SHCWQILSDM LIDQSDYDPY 840
841 TTIWNNHPMG IKWFIKEAIV YFERQQNLQM LAMLCCVILS ARRKKIPARY YGQELENMEG 900
901 TIVFNDNESQ NTSFWKGSDA FSTRSRSSTV TPNFYGNHLR GKNIHGGDNS SIRSDDHHAR 960
961 LRTHNTLNGS SKFTEPAQKQ GSRAISSSPF HSRMPDIKVE LLHDDIIEAY EQEDLLHLEV1020
1021 SDIPKFQTYI YQYSKLLFRW GLPLERVKIL KVSTDFRSSY SSQGIPPNNN KKSPYNGVLT1080
1081 HWIENNEFGE EKFLARNCNY CDLRVTRSSF ICGNCQHVLH SSCARIWWEI GDECPSGCGC1140
1141 NCPEMFDA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
CMR2 MTC5 |
| View Details | Ho Y, et al. (2002) |
|
AAC3 APT1 ARO1 CCT6 CDC33 CPR6 CYS4 DSS1 ECM10 FMP10 HEF3 HOR2 HXT7 MDJ1 MTC5 NPR2 OYE2 PGM2 POX1 PRE1 PST2 PUP3 RFA3 RHR2 RNQ1 SEH1 SOD2 THI21 TMA29 TPS1 TSA2 YHR033W YJR141W YKU80 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
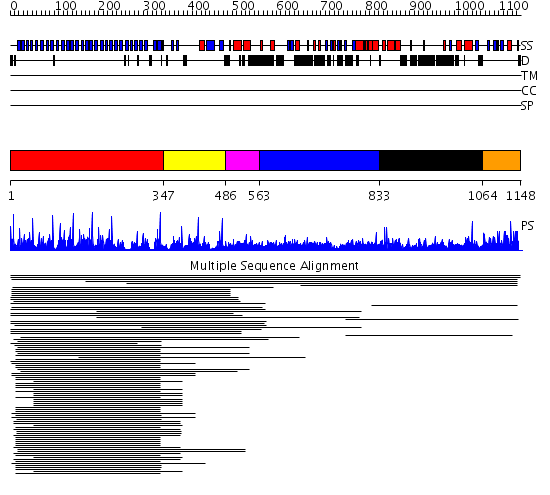
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..346] | 74.0 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 2 | View Details | [347..485] | 2.701996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [486..562] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [563..832] | 1.032964 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [833..1063] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1064..1148] | 1.010996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [968..1062] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1063..1148] | 2.05 | No description for 2d8tA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)