













| General Information: |
|
| Name(s) found: |
RAD51 /
YER095W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 400 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromosome
[IDA condensed nuclear chromosome [IDA |
| Biological Process: |
meiotic joint molecule formation
[IMP reciprocal meiotic recombination [IMP heteroduplex formation [IDA [NOT]double-strand break repair via single-strand annealing [IMP telomere maintenance via recombination [IMP strand invasion [IDA |
| Molecular Function: |
DNA-dependent ATPase activity
[IDA recombinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQVQEQHIS ESQLQYGNGS LMSTVPADLS QSVVDGNGNG SSEDIEATNG SGDGGGLQEQ 60
61 AEAQGEMEDE AYDEAALGSF VPIEKLQVNG ITMADVKKLR ESGLHTAEAV AYAPRKDLLE 120
121 IKGISEAKAD KLLNEAARLV PMGFVTAADF HMRRSELICL TTGSKNLDTL LGGGVETGSI 180
181 TELFGEFRTG KSQLCHTLAV TCQIPLDIGG GEGKCLYIDT EGTFRPVRLV SIAQRFGLDP 240
241 DDALNNVAYA RAYNADHQLR LLDAAAQMMS ESRFSLIVVD SVMALYRTDF SGRGELSARQ 300
301 MHLAKFMRAL QRLADQFGVA VVVTNQVVAQ VDGGMAFNPD PKKPIGGNIM AHSSTTRLGF 360
361 KKGKGCQRLC KVVDSPCLPE AECVFAIYED GVGDPREEDE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
RAD51 RAD52 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
RAD51 RAD52 RAD54 RAD55 RAD57 |
| View Details | Krogan NJ, et al. (2006) |

|
RAD51 RAD52 |
| View Details | Gavin AC, et al. (2006) |
|
ARO1 HMG1 RAD51 RPL27B YHB1 |
| View Details | Gavin AC, et al. (2002) |
|
ARO1 BSP1 BZZ1 CCT8 CHC1 CLU1 CPA2 ECM25 END3 GCN20 GFA1 GUS1 HSC82 IDH2 ILV1 INP52 KAP123 LAS17 LSB3 MKT1 PFK1 RAD51 RAD52 RFA1 RPA135 RPB2 RPN9 RPT3 RPT6 RVB2 SLA1 SLA2 STM1 SYP1 TFP1 VRP1 YHB1 YPT7 YSC84 |
| View Details | Ho Y, et al. (2002) |

|
MLH1 RAD51 |
| View Details | Qiu et al. (2008) |

|
DMC1 RAD51 RAD52 RAD54 RAD55 RAD57 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
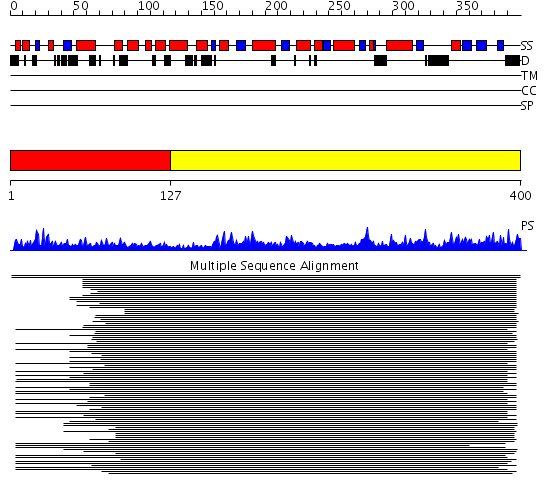
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 311.69897 | DNA repair protein Rad51, N-terminal domain | |
| 2 | View Details | [127..400] | 151.154902 | RecA protein, ATPase-domain; RecA protein, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)