













| General Information: |
|
| Name(s) found: |
MRPS5 /
YBR251W
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 307 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial small ribosomal subunit [IDA |
| Biological Process: |
translation
[IDA |
| Molecular Function: |
structural constituent of ribosome
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFKRQLSTSV RYLQHYDESL LSRYYPESLL KSIKLAQQTI PEDTKFRVSR NVEFAPPYLD 60
61 DFTKIHPFWD YKPGMPHLHA QEENNNFSIF RWDQVQQPLP GEGNILPPGV SLPNDGGRKS 120
121 KSADVAAGLH KQTGVDPDYI TRKLTMKPLV MKRVSNQTGK GKIASFYALV VVGDKNGMVG 180
181 LGEGKSREEM SKAIFKAHWD AVRNLKEIPR YENRTIYGDI DFRYHGVKLH LRSAKPGFGL 240
241 RVNHVIFEIC ECAGIKDLSG KVYKSRNDMN IAKGTIEAFT KAQKTLDEVA LGRGKKLVDV 300
301 RKVYYSS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
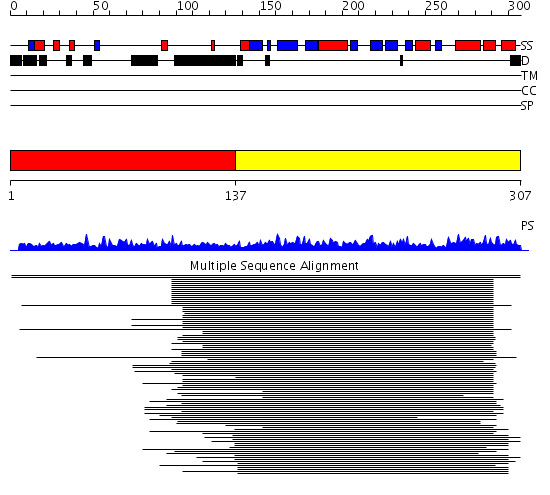
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | 3.128995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [137..307] | 204.9794 | Ribosomal protein S5, C-terminal domain; Ribosomal S5 protein, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)