













| General Information: |
|
| Name(s) found: |
CBF1 /
YJR060W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 351 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA kinetochore [TAS |
| Biological Process: |
methionine biosynthetic process
[IMP response to drug [IMP chromatin assembly or disassembly [IDA chromosome segregation [IMP |
| Molecular Function: |
DNA binding
[IDA centromeric DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSLANNNKL STEDEEIHSA RKRGYNEEQN YSEARKKQRD QGLLSQESND GNIDSALLSE 60
61 GATLKGTQSQ YESGLTSNKD EKGSDDEDAS VAEAAVAATV NYTDLIQGQE DSSDAHTSNQ 120
121 TNANGEHKDS LNGERAITPS NEGVKPNTSL EGMTSSPMES TQQSKNDMLI PLAEHDRGPE 180
181 HQQDDEDNDD ADIDLKKDIS MQPGRRGRKP TTLATTDEWK KQRKDSHKEV ERRRRENINT 240
241 AINVLSDLLP VRESSKAAIL ACAAEYIQKL KETDEANIEK WTLQKLLSEQ NASQLASANE 300
301 KLQEELGNAY KEIEYMKRVL RKEGIEYEDM HTHKKQENER KSTRSDNPHE A |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CBF1 CSE4 MET28 MET4 MIF2 |
| View Details | Krogan NJ, et al. (2006) |

|
CBF1 FYV4 MAM33 MRP1 MRP10 MRP17 MRP21 MRP4 MRP51 MRPL8 MRPS12 MRPS16 MRPS17 MRPS18 MRPS28 MRPS35 MRPS5 MRPS9 NAM9 NCL1 PET123 RIB4 RSM10 RSM18 RSM19 RSM22 RSM23 RSM24 RSM25 RSM26 RSM27 RSM28 RSM7 SVL3 SWS2 YDR124W YJR096W YKL151C YSP1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
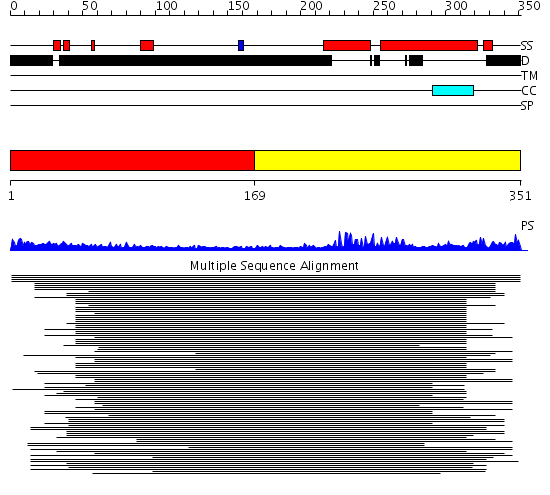
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [169..351] | 11.522879 | Max protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)