













| General Information: |
|
| Name(s) found: |
SMC1 /
YFL008W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1225 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear cohesin complex
[IDA |
| Biological Process: |
mitotic sister chromatid cohesion
[IMP mitotic sister chromatid segregation [IMP double-strand break repair [IMP |
| Molecular Function: |
ATPase activity
[ISS double-stranded DNA binding [IDA DNA secondary structure binding [IDA AT DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGRLVGLELS NFKSYRGVTK VGFGESNFTS IIGPNGSGKS NMMDAISFVL GVRSNHLRSN 60
61 ILKDLIYRGV LNDENSDDYD NEGAASSNPQ SAYVKAFYQK GNKLVELMRI ISRNGDTSYK 120
121 IDGKTVSYKD YSIFLENENI LIKAKNFLVF QGDVEQIAAQ SPVELSRMFE EVSGSIQYKK 180
181 EYEELKEKIE KLSKSATESI KNRRRIHGEL KTYKEGINKN EEYRKQLDKK NELQKFQALW 240
241 QLYHLEQQKE ELTDKLSALN SEISSLKGKI NNEMKSLQRS KSSFVKESAV ISKQKSKLDY 300
301 IFKDKEKLVS DLRLIKVPQQ AAGKRISHIE KRIESLQKDL QRQKTYVERF ETQLKVVTRS 360
361 KEAFEEEIKQ SARNYDKFKL NENDLKTYNC LHEKYLTEGG SILEEKIAVL NNDKREIQEE 420
421 LERFNKRADI SKRRITEELS ITGEKLDTQL NDLRVSLNEK NALHTERLHE LKKLQSDIES 480
481 ANNQEYDLNF KLRETLVKID DLSANQRETM KERKLRENIA MLKRFFPGVK GLVHDLCHPK 540
541 KEKYGLAVST ILGKNFDSVI VENLTVAQEC IAFLKKQRAG TASFIPLDTI ETELPTLSLP 600
601 DSQDYILSIN AIDYEPEYEK AMQYVCGDSI ICNTLNIAKD LKWKKGIRGK LVTIEGALIH 660
661 KAGLMTGGIS GDANNRWDKE EYQSLMSLKD KLLIQIDELS NGQRSNSIRA REVENSVSLL 720
721 NSDIANLRTQ VTQQKRSLDE NRLEIKYHND LIEKEIQPKI TELKKKLDDL ENTKDNLVKE 780
781 KEALQNNIFK EFTSKIGFTI KEYENHSGEL MRQQSKELQQ LQKQILTVEN KLQFETDRLS 840
841 TTQRRYEKAQ KDLENAQVEM KSLEEQEYAI EMKIGSIESK LEEHKNHLDE LQKKFVTKQS 900
901 ELNSSEDILE DMNSNLQVLK RERDGIKEDI EKFDLERVTA LKNCKISNIN IPISSETTID 960
961 DLPISSTDNE AITISNSIDI NYKGLPKKYK ENNTDSARKE LEQKIHEVEE ILNELQPNAR1020
1021 ALERYDEAEG RFEVINNETE QLKAEEKKIL NQFLKIKKKR KELFEKTFDY VSDHLDAIYR1080
1081 ELTKNPNSNV ELAGGNASLT IEDEDEPFNA GIKYHATPPL KRFKDMEYLS GGEKTVAALA1140
1141 LLFAINSYQP SPFFVLDEVD AALDITNVQR IAAYIRRHRN PDLQFIVISL KNTMFEKSDA1200
1201 LVGVYRQQQE NSSKIITLDL SNYAE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
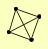
|
IRR1 MCD1 SMC1 SMC3 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
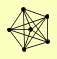
|
IRR1 MCD1 REC8 SCC2 SMC1 SMC3 |
| View Details | Krogan NJ, et al. (2006) |
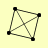
|
IRR1 MCD1 SMC1 SMC3 |
| View Details | Gavin AC, et al. (2006) |

|
SMC1 SMC3 |
| View Details | Gavin AC, et al. (2002) |
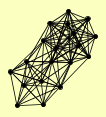
|
ACT1 CCT8 ENO2 FPR4 GCN1 HHF1, HHF2 IRR1 KAP123 MCD1 NAN1 NOP12 POL5 PWP1 RVB2 SCC4 SMC1 SMC3 TYW1 YHB1 |
| View Details | Ho Y, et al. (2002) |
|
CDC5 IRR1 IVY1 KAP95 MCD1 NOP13 SMC1 SMC3 SPC72 SRP1 |
| View Details | Qiu et al. (2008) |

|
SCC2 SMC1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
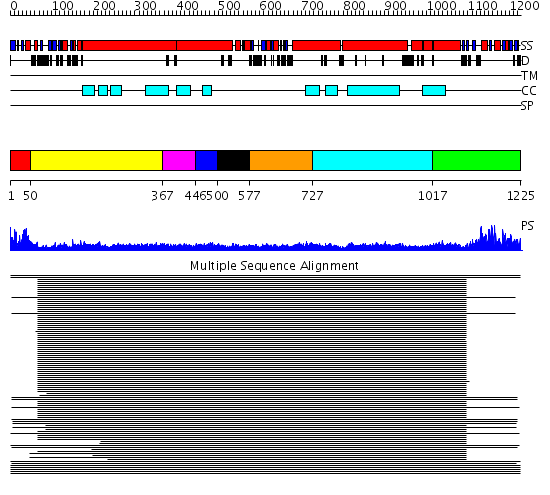
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..49] | 10.32 | Rad50 | |
| 2 | View Details | [50..366] | 27.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [367..445] | 9.221849 | Tropomyosin | |
| 4 | View Details | [446..499] | 9.221849 | Tropomyosin | |
| 5 | View Details | [500..576] | 134.0 | Smc hinge domain | |
| 6 | View Details | [577..726] | 5.30103 | Colicin Ia; Colicin Ia, N-terminal domain | |
| 7 | View Details | [727..1016] | 5.30103 | Colicin Ia; Colicin Ia, N-terminal domain | |
| 8 | View Details | [1017..1225] | 167.519517 | Smc head domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)