













| General Information: |
|
| Name(s) found: |
lola-PP /
FBpp0088389
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 963 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
|
| Biological Process: |
regulation of gene-specific transcription
[IMP]
R7 cell development [IMP] axon guidance [IGI][IMP][NAS] nurse cell apoptosis [IMP] brain morphogenesis [IMP] R3/R4 cell fate commitment [IMP] inter-male aggressive behavior [IMP] regulation of transcription from RNA polymerase II promoter [NAS] locomotion involved in locomotory behavior [IMP] startle response [IMP] positive regulation of transcription, DNA-dependent [ISS] axonogenesis [IMP] axon midline choice point recognition [IGI][IMP] antimicrobial humoral response [IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IPI] RNA polymerase II transcription factor activity [NAS] specific RNA polymerase II transcription factor activity [ISS][NAS] transcription activator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDDQQFCLR WNNHQSTLIS VFDTLLENET LVDCTLAAEG KFLKAHKVVL SACSPYFATL 60
61 LQEQYDKHPI FILKDVKYQE LRAMMDYMYR GEVNISQDQL AALLKAAESL QIKGLSDNRT 120
121 GGGVAPKPES SGHHRGGKLS GAYTLEQTKR ARLATGGAMD TSGDVSGSRE GSSSPSRRRR 180
181 KVRRRSMEND AHDNSNSSVL QAAASNQSIL QQTGAGLAVS ALVTTQLSSG PAAGTSSQAS 240
241 STQQQQPLTS TNVTKKTESA KLTSSTAAPA SGASASAAVQ QAHLHQQQAQ TTSDAINTEN 300
301 VQAQSQGGAQ GVQGDDEDID EGSAVGGPNS ATGPNPASAS ASAVHAGVVV KQLASVVDKS 360
361 SSNHKHKIKD NSVSSVGSEM VIEPKAEYDD DAHDENVEDL TLDEEDMTME ELDQTAGTSQ 420
421 GGEGSSQTYA TWQHDRSQDE LGLMAQDAQQ RDPQDEAGQN EGGESRIRVR NWLMLADKSI 480
481 IGKSSDEPSD KLTQSKKSLI SDAKTTNKTS TPIRPKVSTT TTSTSTAAAA AAAATIAAKQ 540
541 AAAAIASSNI NNNNSSLTQT VTQTVTRIGS IGRTTIACIT PANNGNKSSS SNCNVDAASA 600
601 AALAAAGVEL DSIDDTMTEV IVKIENPESM PLNDDEDDAV CNEAIEDENT FDYDLKLGSP 660
661 LSWTYDAVKI ENEEFEDSYL MDNDDDDDDL LTTAAATQKH AKQSNEKQMA GSMVAGAGSG 720
721 GAVKKIVLSA QQQQQLLEQQ QHLQHLQLQP TSQSLQIKLP AIPATITTIS APKQMMSGAG 780
781 TSGSLTPNNN CTLMSNKLGL PVKGQNLDLH WSHSDDNRYR VLVQNKRTRK ESLEHSADMI 840
841 YNADIEKPWV CRNCNRTYKW KNSLKCHLKN ECGLPPRYFC SKMCGYATNV HSNLKRHLNT 900
901 KCRDREKDAD DEKKPGSASG NMPVVVGVGN GTAVPVSSSN NNNNGGGSST SSTYTLVFQN 960
961 DSA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
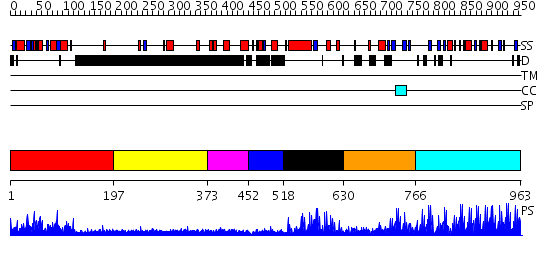
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 33.30103 | Crystal Structure of the B-Cell Lymphoma 6 (BCL6) BTB domain to 2.2 Angstrom | |
| 2 | View Details | [197..372] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [373..451] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [452..517] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [518..629] | 3.69897 | No description for 2dlkA was found. | |
| 6 | View Details | [630..765] | 1.729996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [766..963] | 52.09691 | No description for 2i13A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)