













| General Information: |
|
| Name(s) found: |
pdk-1 /
CE28740
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 636 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
neuronal cell body [IDA] axon [IDA] |
| Biological Process: |
dauer larval development
[IGI regulation of chemotaxis [IMP protein amino acid phosphorylation [IEA learning or memory [IMP |
| Molecular Function: |
magnesium ion binding
[IEA ATP binding [IEA protein tyrosine kinase activity [IEA zinc ion binding [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDLTPTNTS LDTTTTNNDT TSDREAAPTT LNLTPTASES ENSLSPVTAE DLIAKSIKEG 60
61 CPKRTSNDFM FLQSMGEGAY SQVFRCREVA TDAMFAVKVL QKSYLNRHQK MDAIIREKNI 120
121 LTYLSQECGG HPFVTQLYTH FHDQARIYFV IGLVENGDLG ESLCHFGSFD MLTSKFFASE 180
181 ILTGLQFLHD NKIVHRDMKP DNVLIQKDGH ILITDFGSAQ AFGGLQLSQE GFTDANQASS 240
241 RSSDSGSPPP TRFYSDEEVP EENTARRTTF VGTALYVSPE MLADGDVGPQ TDIWGLGCIL 300
301 FQCLAGQPPF RAVNQYHLLK RIQELDFSFP EGFPEEASEI IAKILVRDPS TRITSQELMA 360
361 HKFFENVDWV NIANIKPPVL HAYIPATFGE PEYYSNIGPV EPGLDDRALF RLMNLGNDAS 420
421 ASQPSTFRPS NVEHRGDPFV SEIAPRANSE AEKNRAARAQ KLEEQRVKNP FHIFTNNSLI 480
481 LKQGYLEKKR GLFARRRMFL LTEGPHLLYI DVPNLVLKGE VPWTPCMQVE LKNSGTFFIH 540
541 TPNRVYYLFD LEKKADEWCK AINDVRKRYS VTIEKTFNSA MRDGTFGSIY GKKKSRKEMM 600
601 REQKALRRKQ EKEEKKALKA EQVSKKLSMQ MDKKSP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
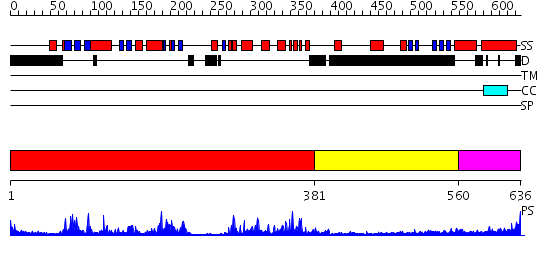
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..380] | 66.09691 | Structure of human PDK1 kinase domain in complex with staurosporine | |
| 2 | View Details | [381..559] | 21.30103 | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain bound to Inositol (1,3,4,5)-tetrakisphosphate | |
| 3 | View Details | [560..636] | 4.0 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)