













| General Information: |
|
| Name(s) found: |
CalpC-PA /
FBpp0074005
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 681 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[NAS]
intracellular [IEA] |
| Biological Process: |
proteolysis
[IEA][NAS]
|
| Molecular Function: |
calcium ion binding
[IEA]
[NOT]calcium-dependent cysteine-type endopeptidase activity [IEA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASKYERILS DCRSKNVLWE DPDFPAVQSS VFYYQTPPFT FQWKRIMDLA DSGSGAVAAN 60
61 SSAAPVFLNE SAEFDVVPGK MGDRWLVSCL GLLSSLRNLF YRVVPADQTL ASAHGVFRFR 120
121 LWWCGEWVEV LVDDRLPTIN GRLAFMQPQA SNCFWAALLE KAIAKLHGSY EALKYGTRSD 180
181 GLTDLLGGVV RQMPILSDNI RPQTLKELLT TTCIVTCLAD KSATVAKKNL AERMPNGILV 240
241 NVNYRLSSLD KVKTLMGDSV QLVCLKDTFS SKPFGEKTHF LGDWSPMSKT WERVSQVERA 300
301 RLIRQLGPGE FWLSFCDFVE IFSTMEVVYL DTETSNDEEM LKSRPLHWKM KMHQGQWKRG 360
361 VTAGGCRNHE SFHINPQLLI SVQDEQDLVI ALNQHTAVEP KVIGFTMYTW DGEYMLSECL 420
421 QKDFFKNHVS YLNSDYGNTR HVSYHTHLEA GHYVLIPTTY EPAEEAHFTV RILGTGSFRL 480
481 SCLETQTMIL LDPFPALKST DAERCGGPKV KSVCQYEPVY MQLADENKTI NCFELHELLE 540
541 ACLPNDYIKG CANIDICRQV IALQDRSGSG RITFQQFKTF MVNLKSWQGV FKMYTKEKAG 600
601 ILRAERLRDA LCDIGFQLST DIMNCLIQRY IRKDGTLRLS DFVSAVIHLT TAFNQFHLKN 660
661 YGQVNVIEVH LHDWIKSILS C |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
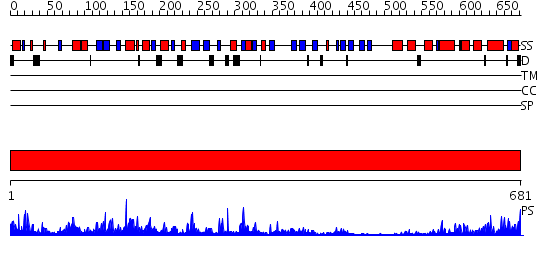
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..681] | 165.0 | Calpain large subunit, C-terminal domain (domain IV); Calpain large subunit, middle domain (domain III); Calpain large subunit, catalytic domain (domain II) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)