













| General Information: |
|
| Name(s) found: |
Mcm6-PA /
FBpp0070913
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 817 amino acids |
Gene Ontology: |
|
| Cellular Component: |
pre-replicative complex
[ISS][NAS]
nucleus [IDA] |
| Biological Process: |
DNA replication initiation
[IEA]
eggshell chorion gene amplification [IMP][TAS] oogenesis [TAS] DNA replication [IEA][IMP][TAS] pre-replicative complex assembly [ISS][NAS] |
| Molecular Function: |
ATP binding
[IEA]
DNA binding [IEA] chromatin binding [ISS][NAS] 3'-5' DNA helicase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVADAQVGQ LRVKDEVGIR AQKLFQDFLE EFKEDGEIKY TRPAASLESP DRCTLEVSFE 60
61 DVEKYDQNLA TAIIEEYYHI YPFLCQSVSN YVKDRIGLKT QKDCYVAFTE VPTRHKVRDL 120
121 TTSKIGTLIR ISGQVVRTHP VHPELVSGVF MCLDCQTEIR NVEQQFKFTN PTICRNPVCS 180
181 NRKRFMLDVE KSLFLDFQKI RIQETQAELP RGCIPRAVEI ILRSELVETV QAGDRYDFTG 240
241 TLIVVPDVSV LAGVGTRAEN SSRHKPGEGM DGVTGLKALG MRELNYRMAF LACSVQATTA 300
301 RFGGTDLPMS EVTAEDMKKQ MTDAEWHKIY EMSKDRNLYQ NLISSLFPSI YGNDEVKRGI 360
361 LLQQFGGVAK TTTEKTSLRG DINVCIVGDP STAKSQFLKQ VSDFSPRAIY TSGKASSAAG 420
421 LTAAVVRDEE SFDFVIEAGA LMLADNGICC IDEFDKMDQR DQVAIHEAME QQTISIARAG 480
481 VRATLNARTS ILAAANPING RYDRSKSLQQ NIQLSAPIMS RFDLFFILVD ECNEVVDYAI 540
541 ARKIVDLHSN IEESVERAYT REEVLRYVTF ARQFKPVISQ EAGHMLVENY GHLRQRDTGT 600
601 SGRSTWRITV RQLESMIRLS EAMAKLECSN RVLERHVKEA FRLLNKSIIR VEQPDIHLDD 660
661 DEGLDMDDGI QHDIDMENNG AAANVDENNG MDTSASGAVQ KKKFTLSFED YKNLSTMLVL 720
721 HMRAEEARCE VEGNDTGIKR SNVVTWYLEQ VADQIESEDE LISRKNLIEK LIDRLIYHDQ 780
781 VIIPLKTSTL KPRIQVQKDF VEEDDPLLVV HPNYVVE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
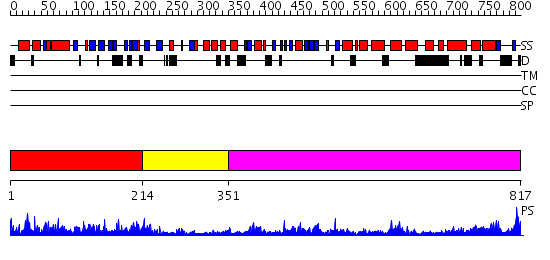
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | 37.522879 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 2 | View Details | [214..350] | 52.154902 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 3 | View Details | [351..817] | 14.09691 | EDTA treated |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)