













| General Information: |
|
| Name(s) found: |
FBpp0309092
[FlyBase]
Klp3A-PA / FBpp0070432 [FlyBase] |
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1212 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
intermediate layer of spindle pole body [IDA] central plaque of spindle pole body [IDA] cytoplasm [IDA] spindle [IDA] aster [IDA] kinesin complex [ISS][NAS] midbody [IDA] microtubule associated complex [IDA] |
| Biological Process: |
reciprocal meiotic recombination
[IMP]
spindle elongation [IMP] regulation of mitosis [IMP] spindle pole body separation [IMP] male meiosis [IMP][TAS] mitotic spindle organization [IMP] pronuclear fusion [TAS] cytokinesis [IMP][TAS] chromosome segregation [IMP] female meiosis chromosome segregation [IMP] microtubule-based movement [ISS] single fertilization [IMP] |
| Molecular Function: |
ATP binding
[IEA]
microtubule motor activity [ISS] plus-end-directed microtubule motor activity [NAS] microtubule binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSEDPSCVA VALRVRPLVQ SELDRGCRIA VERSADGAPQ VTVNRNESYT YNYVFDIDDS 60
61 QKDLFETCVQ AKVKKLLNGY NVTILAYGQT GSGKTYTMGT AFNGVLDDHV GVIPRAVHDI 120
121 FTAIAEMQSE FRFAVTCSFV ELYQEQFYDL FSSKTRDKAT VDIREVKNRI IMPGLTELVV 180
181 TSAQQVTDHL IRGSAGRAVA ATAMNETSSR SHAIFTLTLV ATKLDGKQSV TTSRFNLVDL 240
241 AGSERCSKTL ASGDRFKEGV NINKGLLALG NVINALGSGQ AAGYIPYRQS KLTRLLQDSL 300
301 GGNSITLMIA CVSPADYNVA ETLSTLRYAD RALQIKNKPV VNLDPHAAEV NMLKDVIQKL 360
361 RVELLSGGKM SSSLISAVGA AGLGAIPCEE SLAGSMANAA EIQRLKEQVR TLQDRNRKLQ 420
421 QELHQSLLDL TEKEMRAHIA EQAHDKLRSH VSELKNKLDQ REQAQFGNEN TNGDNEMRDF 480
481 SLLVNRVHVE LQRTQEELES QGHESRQRLS SRSHTEGGES GGDEVHEMLH SHSEEYTNKQ 540
541 MNFAGELRNI NRQLDLKQEL HERIMRNFSR LDSDDEDVKL RLCNQKIDDL EAERRDLMDQ 600
601 LRNIKSKDIS AKLAEERRKR LQLLEQEISD LRRKLITQAN LLKIRDKERE KIQNLSTEIR 660
661 TMKESKVKLI RAMRGESEKF RQWKMVREKE LTQLKSKDRK MQSEIVRQQT LHSKQRQVLK 720
721 RKCEEALAAN KRLKDALERQ ASAQAQRHKY KDNGGSAAGS SNANAKTDSW VDRELEIILS 780
781 LIDAEHSLEQ LMEDRAVINN HYHLLQQEKT SDPAEAAEQA RILASLEEEL EMRNAQISDL 840
841 QQKVCPTDLD SRIRSLAEGV QSLGESRTVS KQLLKTLVQQ RRLQASSLNE QRTTLDELRA 900
901 QLLDAQQQED AASKRLRLLQ SQHEEQMLAQ QRAYEEKVSV LIRTANQRWA EARSPAEDQQ 960
961 RNQILEELLS SREALQQELD KLRAKNKSKS KAVKSEPQDL DDSFQIVDGN ETVVLSDVSD1020
1021 DPDWVPSTSK SKRIQSDSRN VISPPEKQDA NVTSLGNSSI QSLNSTSATE DGKRCKGCKC1080
1081 RTKCTTKRCG CLSGNNACSE TCVCKSNCRN PLNLKDHASQ CGDGDGQKDE TEDADKSDDD1140
1141 GDDEPQTSKE NAVKFVTPEA PGKVVASPKQ TLQEPKAAAT PLMNSNVVED INGPKLAKMS1200
1201 GLAFDTPKRK FF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
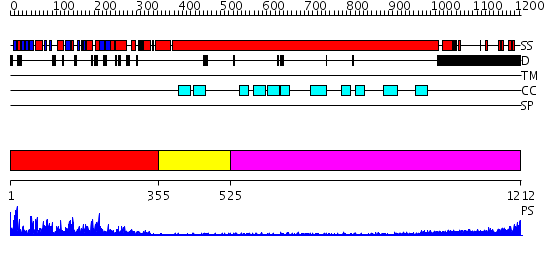
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..354] | 100.0 | Kinesin | |
| 2 | View Details | [355..524] | 1.59 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 3 | View Details | [525..1212] | 5.39794 | No description for 2dfsA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)