













| General Information: |
|
| Name(s) found: |
FBpp0306705
[FlyBase]
Pi3K92E-PB / FBpp0083349 [FlyBase] Pi3K92E-PA / FBpp0083348 [FlyBase] |
| Description(s) found:
Found 49 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1088 amino acids |
Gene Ontology: |
|
| Cellular Component: |
1-phosphatidylinositol-4-phosphate 3-kinase, class IA complex
[IPI][NAS]
|
| Biological Process: |
positive regulation of growth
[TAS]
regulation of multicellular organism growth [IMP][NAS] germ-band shortening [IMP] positive regulation of translation [IMP] insulin receptor signaling pathway [NAS][TAS] regulation of cell size [IMP][NAS] phosphoinositide phosphorylation [IDA][ISS][TAS] metamorphosis [IMP] larval development [IMP] larval salivary gland morphogenesis [IMP] instar larval development [IMP] regulation of dendrite development [IMP] response to oxidative stress [IMP] positive regulation of cell growth [NAS][TAS] positive regulation of cell size [TAS] autophagy [IMP] positive regulation of imaginal disc growth [IMP] phosphorylation [NAS] positive regulation of organ growth [IMP][NAS] regulation of organ growth [NAS] regulation of cell proliferation [IMP] larval feeding behavior [IMP] phosphoinositide-mediated signaling [IEA] oogenesis [IMP] diapause [IMP] cell growth [TAS] |
| Molecular Function: |
lipid kinase activity
[IDA]
phosphatidylinositol-4,5-bisphosphate 3-kinase activity [IDA][ISS] phosphoinositide 3-kinase activity [ISS] binding [IEA] phosphatidylinositol-4-phosphate 3-kinase activity [IDA][ISS] 1-phosphatidylinositol-3-kinase activity [IDA][ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNMMDNRALA YVAHQPKYET PPEEAEPPCM RFSVNLWKNE MLNWVDLICL LPNGFLLELR 60
61 VNPANTIQVI KVEMVNQAKQ MPLGYVIKEA CEYQVYGIST FNIEPYTDET KRLSEVQPYF 120
121 GILSLGERTD TTSFSSDYEL TKMVNGMIGT TFDHNRTHGS PEIDDFRLYM TQTCDNIELE 180
181 RSAYTWQQRL LYEHPLRLAN STKMPELIRE RHPTRTFLIV VKNENDQSTF TLSVNEQDTP 240
241 FSLTESTLQK MNRSQMKMND RTSDYILKVS GRDEYLLGDY PLIQFLYIQE MLSDSAVPNV 300
301 VLQSVYRLES YINHHNEQAM VTKRPLPKKR TVHLHKSISS LWDMGNYFQL TLHSISNVNF 360
361 DKTRALKVGV HVCLYHGDKK LCAQRSTDSP NGNFDTFLFN DLVMDFDIQM RNLPRMTRLC 420
421 IVIFEVTKMS RSKKSSNNKD IALKDVPYNK NPLAWVNTTI FDHKDILRTG RHTLYTWTYA 480
481 DDIQSVEVFH PLGTIEPNPR KEECALVDLT FLSSGTGTVR YPSEEVVLQY AADREQVNRL 540
541 QRQLAGPEKP IKELKELMAN YTGLDKIYEM VDQDRNAIWE RRNDILRELP EELSILLHCV 600
601 YWKERDDVAD MWYLLKQWPL ISIERSLELL DYAYPDPAVR RFAIRCLHFL KDEDLLLYLL 660
661 QLVQAIKHES YLESDLVVFL LERALRNQRI GHYFFWHLRS EMQTPSMQTR FGLLLEVYLK 720
721 GCKHHVAPLR KQLHVLEKLK QGSLIAKKGS KEKVKTMLQD FLRDQRNSAV FQNIQNPLNP 780
781 SFRCSGVTPD RCKVMDSKMR PLWVVFENAD VNASDVHIIF KNGDDLRQDM LTLQMLRVMD 840
841 QLWKRDGMDF RMNIYNCISM EKSLGMIEVV RHAETIANIQ KEKGMFSATS PFKKGSLLSW 900
901 LKEHNKPADK LNKAINEFTL SCAGYCVATY VLGVADRHSD NIMVKRNGQL FHIDFGHILG 960
961 HFKEKLGVRR ERVPFVLTHD FVYVINKGFN DRESKEFCHF QELCERAFLV LRKHGCLILS1020
1021 LFSMMISTGL PELSSEKDLD YLRETLVLDY TEEKAREHFR AKFSEALANS WKTSLNWASH1080
1081 NFSKNNKQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
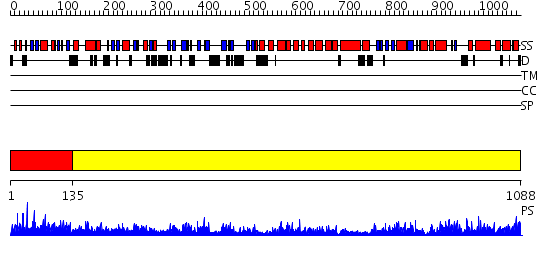
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 31.0 | No description for 2v1yA was found. | |
| 2 | View Details | [135..1088] | 1000.0 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)