













| General Information: |
|
| Name(s) found: |
CG7747-PA /
FBpp0086323
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 517 amino acids |
Gene Ontology: |
|
| Cellular Component: |
catalytic step 2 spliceosome
[IDA]
|
| Biological Process: |
protein folding
[IEA]
nuclear mRNA splicing, via spliceosome [IC] |
| Molecular Function: |
peptidyl-prolyl cis-trans isomerase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKRQHQKDK MYLTYTEWSE LYGGKKVESL ENDHVKFKRL PFEHCCITMA PYEMPYCDLQ 60
61 GNVFEYEAIL KFLKTFKVNP ITGQKMDSKS LVKLNFHRNA NDEYHCPALF KPFSKNSHIV 120
121 AVATTGNVYC WEAIDQLNIK TKNWKDLVDD TPFQRKDIIT IQDPQKLEKY DISTFYHIKK 180
181 NLRVLTEEEQ QERKNPASGR IKTMNLETKE TLEQLQQDYQ PAEEEASTSK RTADKFNAAH 240
241 YSTGAVAASF TSTAMVPVSQ IEAAIIDDDL VKYERVKKKG YVRLNTNLGP LNLELFCDQT 300
301 PRACDNFIKH CANGYYNNVM FHRSIRNFIV QGGDPTGSGS GGESIWGKKF EDEFKPNLTH 360
361 TGRGVLSMAN SGPNTNGSQF FITYRSCKHL DGKHTIFGKL VGGLDTLQKM ENIEVDNKDR 420
421 PIEDIIIESS QVFVNPFAEA AEQLAKEREE EAAGKEEIVK KEEQQKRMKE PLKVYREGVG 480
481 KYLKLQTVAK KPEAPLTSAQ AAKKKKLANG FGDFSSW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
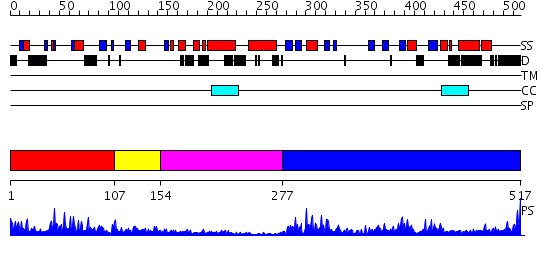
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 14.69897 | Crystal structure of the CHIP U-box E3 ubiquitin ligase | |
| 2 | View Details | [107..153] | 2.02 | Crystal structure of the Prp19 U-box dimer | |
| 3 | View Details | [154..276] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [277..517] | 53.30103 | Crystal Structure of Recombinant Human Cyclophilin J |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)