













| General Information: |
|
| Name(s) found: |
Iswi-PC /
FBpp0086956
[FlyBase]
Iswi-PB / FBpp0086955 [FlyBase] Iswi-PA / FBpp0086954 [FlyBase] |
| Description(s) found:
Found 51 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1027 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription factor complex
[IPI]
ACF complex [IDA][NAS][TAS] [NOT]brahma complex [IDA] chromatin remodeling complex [ISS] chromatin accessibility complex [IDA][NAS][TAS] NURF complex [IDA][NAS][TAS] |
| Biological Process: |
transcription
[IDA]
negative regulation of gene-specific transcription [IMP] ecdysone receptor-mediated signaling pathway [IGI] regulation of transcription, DNA-dependent [IDA] chromatin assembly or disassembly [IDA][TAS] positive regulation of transcription [TAS] regulation of circadian rhythm [IMP] nucleosome assembly [IDA] nucleosome mobilization [IDA][TAS] dendrite morphogenesis [IMP] muscle organ development [IMP] chromatin remodeling [NAS][TAS] ATP-dependent chromatin remodeling [IEA] nucleosome positioning [IDA] |
| Molecular Function: |
nucleotide binding
[TAS]
ATPase activity [NAS] ATP binding [ISS] DNA binding [IEA] DNA-dependent ATPase activity [IDA][IGI] DNA helicase activity [TAS] protein binding [IPI] nucleosome binding [IEA] general RNA polymerase II transcription factor activity [ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKTDTAAVE ATEENSNETT SDAATSSSGE KEAEFDNKIE ADRSRRFDFL LKQTEIFTHF 60
61 MTNSAKSPTK PKGRPKKIKD KDKEKDVADH RHRKTEQEED EELLAEDSAT KEIFRFDASP 120
121 AYIKSGEMRD YQIRGLNWMI SLYENGINGI LADEMGLGKT LQTISLLGYL KHFKNQAGPH 180
181 IVIVPKSTLQ NWVNEFKKWC PSLRAVCLIG DQDTRNTFIR DVLMPGEWDV CVTSYEMCIR 240
241 EKSVFKKFNW RYLVIDEAHR IKNEKSKLSE ILREFKTANR LLITGTPLQN NLHELWALLN 300
301 FLLPDVFNSS EDFDEWFNTN TCLGDDALIT RLHAVLKPFL LRRLKAEVEK RLKPKKEMKI 360
361 FVGLSKMQRD WYTKVLLKDI DVVNGAGKVE KMRLQNILMQ LRKCTNHPYL FDGAEPGPPY 420
421 TTDTHLVYNS GKMAILDKLL PKLQEQGSRV LIFSQMTRML DILEDYCHWR NYNYCRLDGQ 480
481 TPHEDRNRQI QEFNMDNSAK FLFMLSTRAG GLGINLATAD VVIIYDSDWN PQMDLQAMDR 540
541 AHRIGQKKQV RVFRLITEST VEEKIVERAE VKLRLDKMVI QGGRLVDNRS NQLNKDEMLN 600
601 IIRFGANQVF SSKETDITDE DIDVILERGE AKTAEQKAAL DSLGESSLRT FTMDTNGEAG 660
661 TSSVYQFEGE DWREKQKLNA LGNWIEPPKR ERKANYAVDA YFREALRVSE PKAPKAPRPP 720
721 KQPIVQDFQF FPPRLFELLD QEIYYFRKTV GYKVPKNTEL GSDATKVQRE EQRKIDEAEP 780
781 LTEEEIQEKE NLLSQGFTAW TKRDFNQFIK ANEKYGRDDI DNIAKDVEGK TPEEVIEYNA 840
841 VFWERCTELQ DIERIMGQIE RGEGKIQRRL SIKKALDQKM SRYRAPFHQL RLQYGNNKGK 900
901 NYTEIEDRFL VCMLHKLGFD KENVYEELRA AIRASPQFRF DWFIKSRTAL ELQRRCNTLI 960
961 TLIERENIEL EEKERAEKKK KAPKGSVSAG SGSASSNTPA PAPQPKASQK RKSEVVATSS1020
1021 NSKKKKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
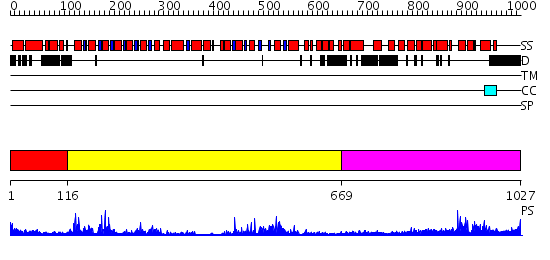
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 4.09691 | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | |
| 2 | View Details | [116..668] | 85.154902 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 3 | View Details | [669..1027] | 59.522879 | nucleosome recognition module of ISWI ATPase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)