













| General Information: |
|
| Name(s) found: |
YTH1 /
YPR107C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 208 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mRNA cleavage and polyadenylation specificity factor complex
[IPI |
| Biological Process: |
mRNA polyadenylation
[IDA mRNA cleavage [IDA |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLIHPDTAK YPFKFEPFLR QEYSFSLDPD RPICEFYNSR EGPKSCPRGP LCPKKHVLPI 60
61 FQNKIVCRHW LRGLCKKNDQ CEYLHEYNLR KMPECVFFSK NGYCTQSPDC QYLHIDPASK 120
121 IPKCENYEMG FCPLGSSCPR RHIKKVFCQR YMTGFCPLGK DECDMEHPQF IIPDEGSKLR 180
181 IKRDDEINTR KMDEEKERRL NAIINGEV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CFT1 CFT2 FIP1 GLC7 MPE1 PAP1 PFS2 PTA1 PTI1 REF2 SSU72 SWD2 YSH1 YTH1 |
| View Details | Krogan NJ, et al. (2006) |

|
AIM1 CFT1 CFT2 ECM21 FIP1 GIP4 MPE1 PAP1 PFS2 PTA1 PTI1 REF2 SSU72 SWD2 SYC1 YBL010C YLR358C YSH1 YTH1 |
| View Details | Gavin AC, et al. (2006) |
|
CFT1 CFT2 FIP1 MPE1 PAP1 PFS2 PTA1 YSH1 YTH1 |
| View Details | Gavin AC, et al. (2002) |

|
ACT1 AIM31 CFT1 CFT2 CKA1 CLP1 ENO2 FIP1 GLC7 HHF1, HHF2 HTA1 MPE1 PAP1 PCF11 PFK1 PFS2 PTA1 PTI1 REF2 RNA14 RNA15 RSA3 SEC13 SEC31 SSA3 SSU72 SYC1 TAF6 TIF4632 TYE7 VID24 VMR1 VPS53 YCL046W YSH1 YTH1 |
| View Details | Qiu et al. (2008) |
|
CFT1 CFT2 CLP1 HRP1 PFS2 PTI1 REF2 RNA14 RNA15 YTH1 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
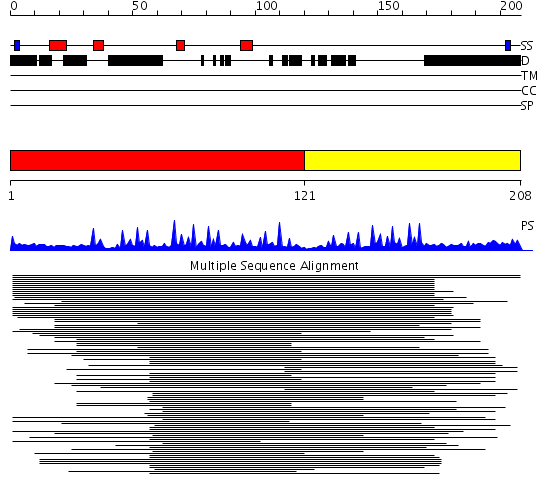
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 6.0 | Tristetraproline (ttp, tis11, nup475) | |
| 2 | View Details | [121..208] | 5.111994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)