













| General Information: |
|
| Name(s) found: |
VPS36 /
YLR417W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ESCRT II complex
[IDA |
| Biological Process: |
protein retention in Golgi apparatus
[IMP ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway [IC telomere maintenance [IMP negative regulation of transcription from RNA polymerase II promoter by glucose [IMP protein targeting to vacuole [IMP |
| Molecular Function: |
regulator of G-protein signaling activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEYWHYVETT SSGQPLLREG EKDIFIDQSV GLYHGKSKIL QRQRGRIFLT SQRIIYIDDA 60
61 KPTQNSLGLE LDDLAYVNYS SGFLTRSPRL ILFFKDPSSK DELGKSAETA SADVVSTWVC 120
121 PICMVSNETQ GEFTKDTLPT PICINCGVPA DYELTKSSIN CSNAIDPNAN PQNQFGVNSE 180
181 NICPACTFAN HPQIGNCEIC GHRLPNASKV RSKLNRLNFH DSRVHIELEK NSLARNKSSH 240
241 SALSSSSSTG SSTEFVQLSF RKSDGVLFSQ ATERALENIL TEKNKHIFNQ NVVSVNGVDM 300
301 RKGASSHEYN NEVPFIETKL SRIGISSLEK SRENQLLNND ILFNNALTDL NKLMSLATSI 360
361 ERLYKNSNIT MKTKTLNLQD ESTVNEPKTR RPLLILDREK FLNKELFLDE IAREIYEFTL 420
421 SEFKDLNSDT NYMIITLVDL YAMYNKSMRI GTGLISPMEM REACERFEHL GLNELKLVKV 480
481 NKRILCVTSE KFDVVKEKLV DLIGDNPGSD LLRLTQILSS NNSKSNWTLG ILMEVLQNCV 540
541 DEGDLLIDKQ LSGIYYYKNS YWPSHI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SNF8 VPS36 |
| View Details | Krogan NJ, et al. (2006) |
|
ADH7 HCM1 OLA1 SNF8 VPS25 VPS36 |
| View Details | Qiu et al. (2008) |
|
DID4 SNF8 VPS20 VPS25 VPS36 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
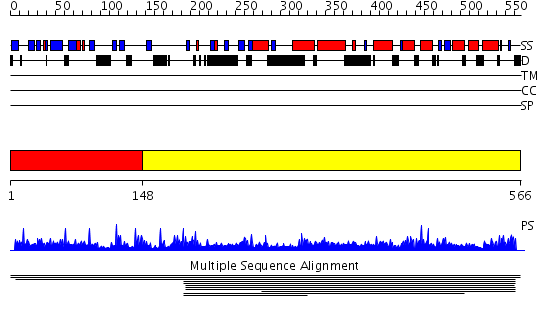
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [148..566] | 3.010891 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)