













| General Information: |
|
| Name(s) found: |
ADH7 /
YCR105W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 361 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[IDA |
| Biological Process: |
alcohol metabolic process
[IDA |
| Molecular Function: |
alcohol dehydrogenase (NADP+) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLYPEKFQGI GISNAKDWKH PKLVSFDPKP FGDHDVDVEI EACGICGSDF HIAVGNWGPV 60
61 PENQILGHEI IGRVVKVGSK CHTGVKIGDR VGVGAQALAC FECERCKSDN EQYCTNDHVL 120
121 TMWTPYKDGY ISQGGFASHV RLHEHFAIQI PENIPSPLAA PLLCGGITVF SPLLRNGCGP 180
181 GKRVGIVGIG GIGHMGILLA KAMGAEVYAF SRGHSKREDS MKLGADHYIA MLEDKGWTEQ 240
241 YSNALDLLVV CSSSLSKVNF DSIVKIMKIG GSIVSIAAPE VNEKLVLKPL GLMGVSISSS 300
301 AIGSRKEIEQ LLKLVSEKNV KIWVEKLPIS EEGVSHAFTR MESGDVKYRF TLVDYDKKFH 360
361 K |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ADH7 HCM1 OLA1 SNF8 VPS25 VPS36 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
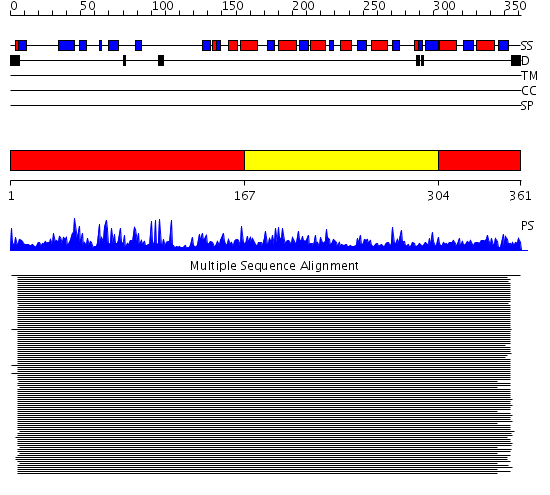
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] [304..361] |
217.9897 | Alcohol dehydrogenase | |
| 2 | View Details | [167..303] | 217.9897 | Alcohol dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.43 |
Source: Reynolds et al. (2008)