













| General Information: |
|
| Name(s) found: |
FRA1 /
YLL029W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 749 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSKPSTSDG RAHSISHVPG THMRGTSASH SPRPFRPCAD CTCSPGLLSR QGRRASLFLR 60
61 QLENSRRSSS MLLNELKGAG GGSSAGNGSV YSCDSLCAVN REVNTTDRLL KLRQEMKKHD 120
121 LCCYIVPSCD EHQSEYVSLR DQRRAFISGF SGSAGVACIT RDLLNFNDDH PDGKSILSTD 180
181 GRYFNQARQE LDYNWTLLRQ NEDPITWQEW CVREALEMAK GLGNKEGMVL KIGIDPKLIT 240
241 FNDYVSFRKM IDTKYDAKGK VELVPVEENL VDSIWPDFET LPERPCNDLL LLKYEFHGEE 300
301 FKDKKEKLLK KLNDKASSAT TGRNTFIVVA LDEICWLLNL RGSDIDYNPV FFSYVAINED 360
361 ETILFTNNPF NDDISEYFKI NGIEVRPYEQ IWEHLTKITS QASSAEHEFL IPDSASWQMV 420
421 RCLNTSTNAN GAIAKKMTAQ NFAIIHSPID VLKSIKNDIE IKNAHKAQVK DAVCLVQYFA 480
481 WLEQQLVGRE ALIDEYRAAE KLTEIRKTQR NFMGNSFETI SSTGSNAAII HYSPPVENSS 540
541 MIDPTKIYLC DSGSQFLEGT TDITRTIHLT KPTKEEMDNY TLVLKGGLAL ERLIFPENTP 600
601 GFNIDAIARQ FLWSRGLDYK HGTGHGIGSF LNVHEGPMGV GFRPHLMNFP LRAGNIISNE 660
661 PGYYKDGEYG IRIESDMLIK KATEKGNFLK FENMTVVPYC RKLINTKLLN EEEKTQINEY 720
721 HARVWRTIVH FLQPQSISYK WLKRETSPL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
FRA1 MET17 SIZ1 |
| View Details | Ho Y, et al. (2002) |
|
FRA1 FRA2 GRX3 GRX4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx21: both fusion and trans-SNARE complex formation take place. Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
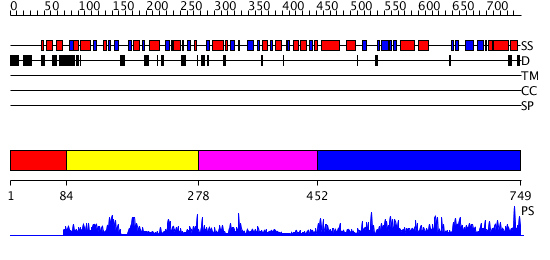
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [84..277] | 8.2 | No description for 1chmA was found. | |
| 3 | View Details | [278..451] | 72.30103 | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | |
| 4 | View Details | [452..749] | 72.30103 | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)