













| General Information: |
|
| Name(s) found: |
RAD26 /
YJR035W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1085 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
transcription-coupled nucleotide-excision repair
[IGI nucleotide-excision repair [IGI |
| Molecular Function: |
DNA-dependent ATPase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDKEQQDNA KLENNESLKD LGVNVLSQSS LEEKIANDVT NFSNLQSLQQ EETRLERSKT 60
61 ALQRYVNKKN HLTRKLNNTT RISVKQNLRD QIKNLQSDDI ERVLKDIDDI QSRIKELKEQ 120
121 VDQGAENKGS KEGLQRPGET EKEFLIRTGK ITAFGHKAGF SLDTANREYA KNDEQKDEDF 180
181 EMATEQMVEN LTDEDDNLSD QDYQMSGKES EDDEEEENDD KILKELEDLR FRGQPGEAKD 240
241 DGDELYYQER LKKWVKQRSC GSQRSSDLPE WRRPHPNIPD AKLNSQFKIP GEIYSLLFNY 300
301 QKTCVQWLYE LYQQNCGGII GDEMGLGKTI QVIAFIAALH HSGLLTGPVL IVCPATVMKQ 360
361 WCNEFQHWWP PLRTVILHSM GSGMASDQKF KMDENDLENL IMNSKPSDFS YEDWKNSTRT 420
421 KKALESSYHL DKLIDKVVTD GHILITTYVG LRIHSDKLLK VKWQYAVLDE GHKIRNPDSE 480
481 ISLTCKKLKT HNRIILSGTP IQNNLTELWS LFDFIFPGKL GTLPVFQQQF VIPINIGGYA 540
541 NATNIQVQTG YKCAVALRDL ISPYLLRRVK ADVAKDLPQK KEMVLFCKLT KYQRSKYLEF 600
601 LHSSDLNQIQ NGKRNVLFGI DILRKICNHP DLLDRDTKRH NPDYGDPKRS GKMQVVKQLL 660
661 LLWHKQGYKA LLFTQSRQML DILEEFISTK DPDLSHLNYL RMDGTTNIKG RQSLVDRFNN 720
721 ESFDVFLLTT RVGGLGVNLT GANRIIIFDP DWNPSTDMQA RERAWRIGQK REVSIYRLMV 780
781 GGSIEEKIYH RQIFKQFLTN RILTDPKQKR FFKIHELHDL FSLGGENGYS TEELNEEVQK 840
841 HTENLKNSKS EESDDFEQLV NLSGVSKLES FYNGKEKKEN SKTEDDRLIE GLLGGESNLE 900
901 TVMSHDSVVN SHAGSSSSNI ITKEASRVAI EAVNALRKSR KKITKQYEIG TPTWTGRFGK 960
961 AGKIRKRDPL KNKLTGSAAI LGNITKSQKE ASKEARQENY DDGITFARSK EINSNTKTLE1020
1021 NIRAYLQKQN NFFSSSVSIL NSIGVSLSDK EDVIKVRALL KTIAQFDKER KGWVLDEEFR1080
1081 NNNAS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
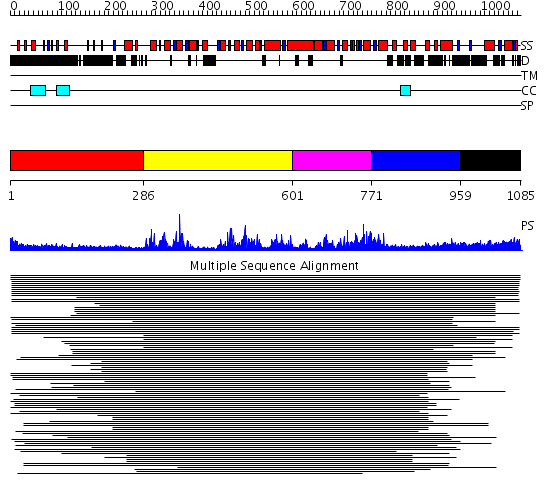
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..285] | 3.300995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [286..600] | 112.79588 | SNF2 family N-terminal domain No confident structure predictions are available. | |
| 3 | View Details | [601..770] | 3.69897 | Putative DEAD box RNA helicase | |
| 4 | View Details | [771..958] | 2.631987 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [959..1085] | 2.051987 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)