













| General Information: |
|
| Name(s) found: |
SRS2 /
YJL092W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1174 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS |
| Biological Process: |
DNA repair
[IGI double-strand break repair via nonhomologous end joining [IDA |
| Molecular Function: |
DNA helicase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSNNDLWLH LVSQLNTQQR AAALFDYTRG LQVIAGPGTG KTKVLTSRVA YLILHHHIHP 60
61 RDIIVTTFTN KAANEMKERL QEMLRGAGVN ISELLIGTFH SICLKILYRF GHLVDLQKDW 120
121 RIIDEKEIDV ILDDMIEKVP DQIRDYASSI TRKVNLCMPS KNGDEWTIHP KLIKKQISKL 180
181 KSNAILPEEY ILDSNHDAAL GYFYQIYQSE LSKKNTLDFD DLLMYTFRLL TRVRVLSNIK 240
241 HVLVDEFQDT NGIQLDLMFL FAKGNHHLSR GMTIVGDPDQ SIYAFRNALA HNFLEMGRKC 300
301 PIEYSTIILV ENYRSSQKIL NTSEILITQQ NKGRQNRAPL RAQFDLDFPP VYMNFPAYFL 360
361 EAPSLVRELL YLKALPNLFT FNDFAILVRQ RRQIKRIESA LIEHRIPYKI IRGHSFWDSK 420
421 ETRAMLNLLK LIFSPNDKHA ILASLLYPAR GLGPATGEKI KNALDTLATD VSCFQILKDI 480
481 SSKKIMLDIP TKGRSVIADF ISMIENCQLL LQSTLLGGLS DLFDKLYELS GLKYEYLYKD 540
541 GKKKNDQLEK SEPNLLNARH KNIELLKNYF LALLSKSESS DKEKNEAIKA ATDEAEPIEN 600
601 KVITPKEYLR NFFNSLSLHS DAAEEEESES NKDAKIKREK NGFVTISTIH GAKGLEWPVV 660
661 FIPGCEEGII PCVFNDDKKD ESEEDEEEDQ ENSKKDASPK KTRVLSVEDS IDEERRMFFV 720
721 AQTRAKYLLY LSNTVTVEDV DRPRIASRFL TTDLIKAMSD SQKLFESTNS IKKLYRILNK 780
781 KPPAEDDKLF SLDQLRKDYN QFIENRRERM IWQGIQMNDV YGIQLSRNKL LGSVSDFTSA 840
841 ADQLRLETQN SIFPQKKLIE KSRPSKINGN YAPKSRVKSP EKRYAPETTS FHSPTKKKVY 900
901 APQYVSTTNV PSRQEFHSST GKNIPFLRRE DRSITDISPR SSTRSLKGAS PNKTSHMSDD 960
961 LMRPSPTRKD KVTRNIHFAT AGTFRIETQS NVDELHPPEY SNKSGQSLTS SEFSGFSSAC1020
1021 SNSDQPTNLI EDINNELDLS DEELLNDISI ERRRELLGSK KTKKIKPKTR NRKSKRGDKV1080
1081 KVEEVIDLKS EFEEDDSRNT TAAELLHNPD DTTVDNRPII SNAKFLADAA MKKTQKFSKK1140
1141 VKNEPASSQM DIFSQLSRAK KKSKLNNGEI IVID |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ACO1 BAT2 HSP42 NTH2 RRP12 SRS2 SSE2 |
| View Details | Ho Y, et al. (2002) |
|
DUN1 SEC23 SEC53 SRS2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
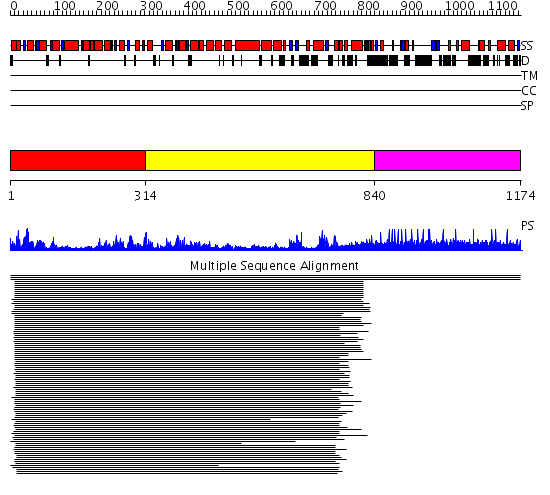
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..313] | 715.9691 | DEXX box DNA helicase | |
| 2 | View Details | [314..839] | 715.9691 | DEXX box DNA helicase | |
| 3 | View Details | [840..1174] | 10.98 | Fibrinogen |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)