













| General Information: |
|
| Name(s) found: |
BAT2 /
YJR148W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 376 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
branched chain family amino acid catabolic process
[IMP branched chain family amino acid biosynthetic process [IDA |
| Molecular Function: |
branched-chain-amino-acid transaminase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLAPLDASK VKITTTQHAS KPKPNSELVF GKSFTDHMLT AEWTAEKGWG TPEIKPYQNL 60
61 SLDPSAVVFH YAFELFEGMK AYRTVDNKIT MFRPDMNMKR MNKSAQRICL PTFDPEELIT 120
121 LIGKLIQQDK CLVPEGKGYS LYIRPTLIGT TAGLGVSTPD RALLYVICCP VGPYYKTGFK 180
181 AVRLEATDYA TRAWPGGCGD KKLGANYAPC VLPQLQAASR GYQQNLWLFG PNNNITEVGT 240
241 MNAFFVFKDS KTGKKELVTA PLDGTILEGV TRDSILNLAK ERLEPSEWTI SERYFTIGEV 300
301 TERSKNGELL EAFGSGTAAI VSPIKEIGWK GEQINIPLLP GEQTGPLAKE VAQWINGIQY 360
361 GETEHGNWSR VVTDLN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
BAT1 BAT2 |
| View Details | Krogan NJ, et al. (2006) |
|
ACO1 BAT2 HSP42 NTH2 RRP12 SRS2 SSE2 |
| View Details | Gavin AC, et al. (2006) |
|
BAT1 BAT2 |
| View Details | Gavin AC, et al. (2002) |

|
BAT1 BAT2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
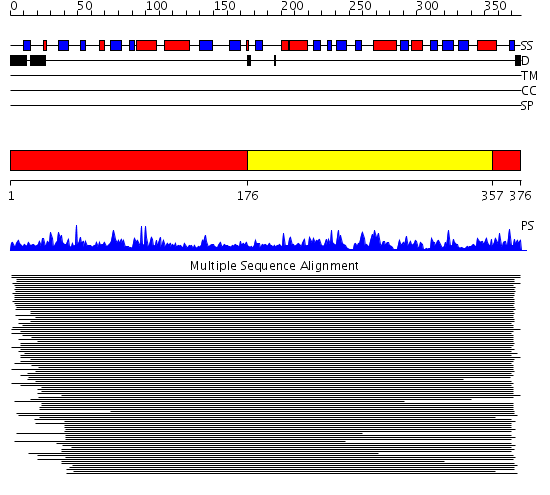
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..175] [357..376] |
978.457575 | Branched-chain aminoacid aminotransferase | |
| 2 | View Details | [176..356] | 978.457575 | Branched-chain aminoacid aminotransferase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)