













| General Information: |
|
| Name(s) found: |
EMI2 /
YDR516C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 500 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
ascospore formation
[IMP response to drug [IMP positive regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFENLHKVN AEALEDAVVE ICSSLQVDAA KLDELTAYFI ECMEKGLNNT SVGEEKTVDK 60
61 GLPMIPTYVT SLPNGTERGV LLAADLGGTH FRVCSVTLNG DGTFDMQQLK SKIPEEYLND 120
121 KDVTSEELFS YLGRRTRAFV RKHHPELLKS TGENIKPLKM GFTFSYPVDQ TSLSSGTLIR 180
181 WTKSFKIEDT VGKDVVRLYQ EQLDIQGLSM INVVALTNDT VGTFLSHCYT SGSRPSSAGE 240
241 ISEPVIGCIF GTGTNGCYME DIENIKKLPD ELRTRLLHEG KTQMCINIEW GSFDNELKHL 300
301 SATKYDIDID QKFSPNPGYH LFEKRISGMY LGELLRNILV DLHARGLILG QYRNYDQLPH 360
361 RLKTPFQLCS EVLSRIEIDD STNLRETELS FLQSLRLPTT FEERKAIQNL VRSITRRSAY 420
421 LAAVPIAAIL IKTNALNKRY HGEVEIGFDG YVIEYYPGFR SMLRHALALS PIGTEGERKI 480
481 HLRLAKDGSG VGAALCALVA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
EMI2 VMA6 VPH2 |
| View Details | Ho Y, et al. (2002) |
|
EMI2 HXT6 SIW14 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
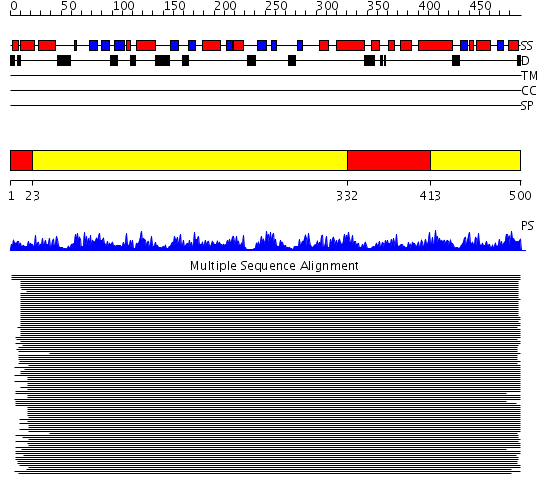
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..22] [332..412] |
824.457575 | Hexokinase | |
| 2 | View Details | [23..331] [413..500] |
824.457575 | Hexokinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)