













| General Information: |
|
| Name(s) found: |
MEK1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 497 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRPLYSCNLA TKDDIEMAGG VAPAHLEVNV GGYNTEQTIP IVKHQLVKVG RNDKECQLVL 60
61 TNPSISSVHC VFWCVFFDED SIPMFYVKDC SLNGTYLNGL LLKRDKTYLL KHCDVIELSQ 120
121 GSEENDIKKT RLVFMINDDL QSSLDPKLLD QMGFLREVDQ WEITNRIVGN GTFGHVLITH 180
181 NSKERDEDVC YHPENYAVKI IKLKPNKFDK EARILLRLDH PNIIKVYHTF CDRNNHLYIF 240
241 QDLIPGGDLF SYLAKGDCLT SMSETESLLI VFQILQALNY LHDQDIVHRD LKLDNILLCT 300
301 PEPCTRIVLA DFGIAKDLNS NKERMHTVVG TPEYCAPEVG FRANRKAYQS FSRAATLEQR 360
361 GYDSKCDLWS LGVITHIMLT GISPFYGDGS ERSIIQNAKI GKLNFKLKQW DIVSDNAKSF 420
421 VKDLLQTDVV KRLNSKQGLK HIWIAKHLSQ LERLYYKKIL CNNEGPKLES INSDWKRKLP 480
481 KSVIISQAIP KKKKVLE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
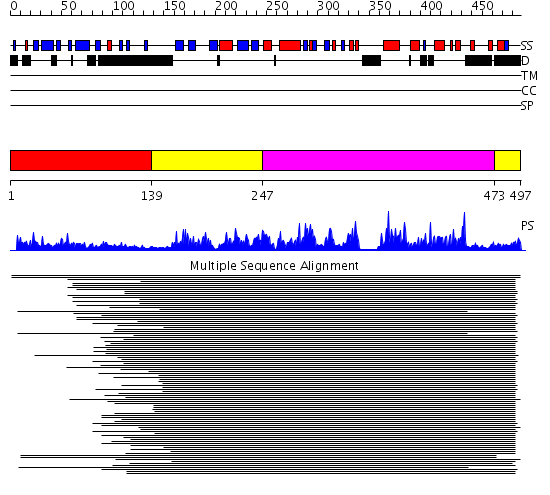
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | 20.39794 | Chk2 kinase | |
| 2 | View Details | [139..246] [473..497] |
475.228787 | Calmodulin-dependent protein kinase | |
| 3 | View Details | [247..472] | 475.228787 | Calmodulin-dependent protein kinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)