













| Protein: | TIF34 |
| From Publication: | Gavin A.C. et al. (2002) Abstract Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002 Jan 10;415(6868):141-7. |
| Complexes containing TIF34: | 2 |
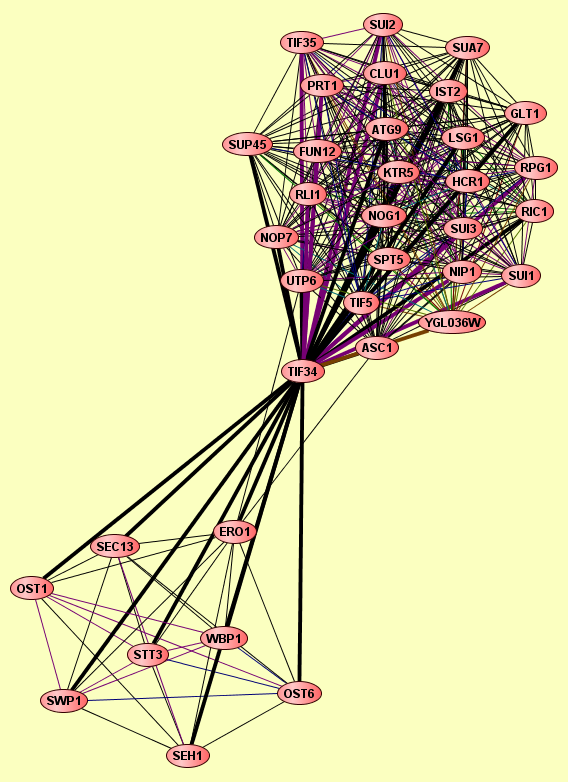
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 9 proteins | From the published set of protein complexes. | ERO1 OST1 OST6 SEC13 SEH1 STT3 SWP1 TIF34 WBP1 |
| View GO Analysis | 27 proteins | From the published set of protein complexes. | ASC1 ATG9 CLU1 FUN12 GLT1 HCR1 IST2 KTR5 LSG1 NIP1 NOG1 NOP7 PRT1 RIC1 RLI1 RPG1 SPT5 SUA7 SUI1 SUI2 SUI3 SUP45 TIF34 TIF35 TIF5 UTP6 YGL036W |