













| General Information: |
|
| Name(s) found: |
gi|9965745
[NCBI NR]
gi|7488072 [NCBI NR] gi|7268139 [NCBI NR] gi|62996992 [NCBI NR] gi|2244791 [NCBI NR] gi|16974431 [NCBI NR] gi|15450525 [NCBI NR] gi|15236471 [NCBI NR] gi|1103318 [NCBI NR] |
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 457 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
phosphorylation
[TAS]
|
| Molecular Function: |
protein serine/threonine kinase activity
[IDA]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDRNQKMDHV IGGKFKLGRK LGSGSFGELY LGINIQTGEE VAVKLEPVKT RHPQLQYESK 60
61 IYMFLQGGTG VPHLKWFGVE GEYSCMVIDL LGPSLEDLFN YCKRIFSLKS VLMLADQLIC 120
121 RVEYMHSRGF LHRDIKPDNF LMGLGRRANQ VYIIDYGLAK KYKDLQTQKH IPYRENKNLT 180
181 GTARYASVNT HLGIEQSRRD DLESLGYVLM YFLRGSLPWQ GLKAGTKKQK YDKISEKKML 240
241 TSVETLCKSY PSEFTSYFHY CRSLRFEDKP DYSYLRRLFR DLFIREGYQL DYVFDWTISK 300
301 YPQIGSSSRP RPTPRPALDP PGPPAERAEK PTVGQDLRGR FTGAIEAFTR RNVSSQGALG 360
361 DRSRHRSSDD IPSSAKEVHE SRNGSTSKRG VISSTRPGSS AEPSENHSSR LFSSGSRHAT 420
421 TQRVPQSYES AAAARPGHED AIRNFELLTI GSGKKRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
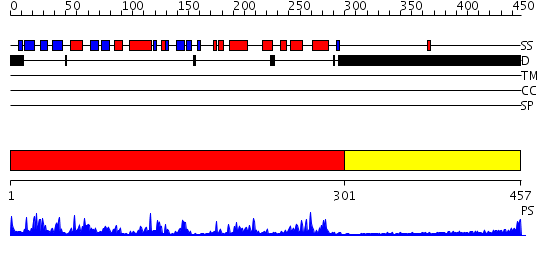
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..300] | 67.522879 | Casein kinase-1, CK1 | |
| 2 | View Details | [301..457] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)