













| General Information: |
|
| Name(s) found: |
osm-3 /
CE31567
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: |
perinuclear region of cytoplasm
[IDA neuronal cell body [IDA neuron projection [IDA microtubule associated complex [IEA microtubule [IDA |
| Biological Process: |
regulation of insulin receptor signaling pathway
[IGI microtubule-based movement [IEA |
| Molecular Function: |
ATPase activity
[ISS ATP binding [IEA microtubule motor activity [IEA protein binding [ISS plus-end-directed microtubule motor activity [ISS microtubule binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPNVGQVNL NAPDGAAKDF TFDGAYFMDS TGEQIYNDIV FPLVENVIEG YNGTVFAYGQ 60
61 TGSGKTFSMQ GIETIPAQRG VIPRAFDHIF TATATTENVK FLVHCSYLEI YNEEVRDLLG 120
121 ADNKQKLEIK EQPDRGVYVA GLSMHVCHDV PACKELMTRG FNNRHVGATL MNKDSSRSHS 180
181 IFTVYVEGMT ETGSIRMGKL NLVDLAGSER QSKTGATGDR LKEATKINLS LSALGNVISA 240
241 LVDGKSKHIP YRDSKLTRLL QDSLGGNTKT IMIACVSPSS DNYDETLSTL RYANRAKNIK 300
301 NKPTINEDPK DALLREYQEE IARLKSMVQP GAVGVGAPAQ DAFSIEEERK KLREEFEEAM 360
361 NDLRGEYERE QTSKAELQKD LESLRADYER ANANLDNLNP EEAAKKIQQL QDQFIGGEEA 420
421 GNTQLKQKRM KQLKEAETKT QKLAAALNVH KDDPLLQVYS TTQEKLDAVT SQLEKEVKKS 480
481 KGYEREIEDL HGEFELDRLD YLDTIRKQDQ QLKLLMQIMD KIQPIIKKDT NYSNVDRIKK 540
541 EAVWNEDESR WILPEMSMSR TILPLANNGY MQEPARQENT LLRSNFDDKL RERLAKSDSE 600
601 NLANSYFKPV KQINVINKYK SDQKLSTSKS LFPSKTPTFD GLVNGVVYTD ALYERAQSAK 660
661 RPPRLASLNP K |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
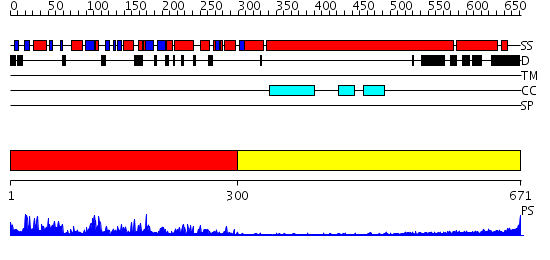
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 84.69897 | Kinesin | |
| 2 | View Details | [300..671] | 4.69897 | No description for 2dfsA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)