













| General Information: |
|
| Name(s) found: |
hsd-2 /
CE07577
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 374 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[IEA |
| Biological Process: |
carbohydrate metabolic process
[IEA steroid biosynthetic process [IEA galactose metabolic process [IEA cellular metabolic process [IEA |
| Molecular Function: |
3-beta-hydroxy-delta5-steroid dehydrogenase activity
[IEA NADP or NADPH binding [IEA coenzyme binding [IEA catalytic activity [IEA UDP-glucose 4-epimerase activity [IEA steroid delta-isomerase activity [IEA ADP-glyceromanno-heptose 6-epimerase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGHYVIVGGG GFLGAHVISA LQKIGCKERI IVVDPCPQEF KTIKIDKSNI SYIKASFLDD 60
61 KVLENILNGA SAVVHLAAVG HTGLIAGDRK SVHNFNVNGT KQLIKQCKAL GVKRFLYASS 120
121 VAVSFIGEPL DNVTEDDPLP DPKKYLDFYS ASKAEAETYV LSQSTPDFKT VCLRFRGIYG 180
181 PEDPNVTLKV ANLIKNGLFI GMVSAHGRES VSCASSGVNC AKAFALADQM LQNPDGLHGR 240
241 AYYILDGENV GQFQFWTPLV LALGKQPPSF YTPYDFIKAV VPYFQNVCYG VFKLPPLLTK 300
301 FELSILAVDN TYSIERARRE LGYEPEPCVM TDVAKYYQDL EENEVATATT EWKSAISILI 360
361 VFISIAFLFL FNLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
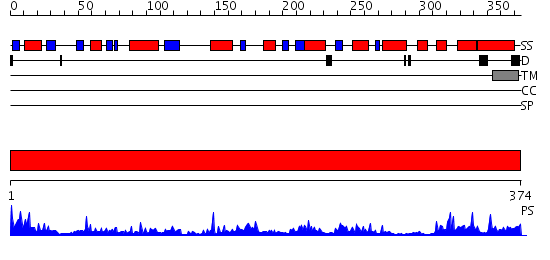
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..374] | 73.045757 | Uridine diphosphogalactose-4-epimerase (UDP-galactose 4-epimerase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|