













| General Information: |
|
| Name(s) found: |
SPBP22H7.05c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1201 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [ISS |
| Biological Process: |
chromatin remodeling
[ISS positive regulation of gene-specific transcription from RNA polymerase II promoter [ISS] |
| Molecular Function: |
ATPase activity
[ISS ATP binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRRRARSIRF SSDDNEDNEE DDDYYSNAHS EKSEDHSNHI KVSHFDPSSY KQKLVSVRET 60
61 QRNRKFSSLQ KHLNTETPSF SVSIENPSKP SAAFNDASLG KKSTEHQIDG IRNGSSNLQM 120
121 EGNDKELDTD NNEDESTTFK DEEDDLISPK SYLTSSKTFS YPKAPTESTN GDYLDEDYVD 180
181 GQSDPESSNA SDSDFADSPD DLTKVRSPIP SRRGRRKRKM RGPILPVKKN LRVKKAMSPL 240
241 RAERNSPDFR RKLRSRDNRP NYHLFDYYNE IASSPNPSTT KITYNPPKLP MKDFATLPIG 300
301 YQSTCDSDET SELSSTSSEQ TSDVEGLNAY NNLGASSDIE NAPSSQLHFG HIDEKTIRST 360
361 DPFANRENLD FNSIGGLEDI ILQLKEMVML PLLYPEVFLH LHITPPRGVL FHGPPGTGKT 420
421 LMARVLAANC STKNQKISFF LRKGSDCLSK WVGEAERQLR LLFEEARRVQ PSIIFFDEID 480
481 GLAPIRSSKQ EQTHSSIVST LLALMDGLDT RGQVVVIGAT NRPNDLDPAL RRPGRFDREF 540
541 YFPLPNKQAR MKILEINSLH FSPKIPESYL LHLAESTSGY GGADLKALCT EAALNAVRRT 600
601 FPQIYTSSDK FLIDLNEISV SICDFVVASE KIAVSTRRSD VKPNIPITDS HKILFKKSIE 660
661 VITSKIRRLL KLDVYLPTVE SLQKLPAEEL MRQKEINSLK TTMSFRPRLL ITDIYGYGCT 720
721 YLSKVLFSML DGIHVQSLDI SELLMDTTTS PRSLLTKIFS EARKNAPSII FINNVEKWPS 780
781 LFSHSFLSMF LLLLDSISPL EPVMLLGFAN TNQEKLSSTV RSWFPSHRSE YHDLSFPDYS 840
841 SRYSFFHYLL KRISFLPIHQ KSAEAASVDI LPKVLPVSKT SDLTDKVNRR QRKNDKKIKN 900
901 KIQVKLSSIL EMLRSRYKKF KKPIIDLNDI YIDESNERVV KGKSKDNFEY FLSGNTVTRK 960
961 KDNACFKMMN FEEIERRLWS GRYCTPKEFL RDIKMIKQDA ILSGDVNLKH KAKEMFAHAE1020
1021 LNVDELIDAK LLYDCCQVSK REKAYKQLKQ KKLNNAKDAH EMQESKNEET FVRNDVAQED1080
1081 NFIELSSNEV RNVSNDEHKH TLFHGQSLTH NNLIAVTPPS RTGVEHKEEN KKYDNVNIQK1140
1141 TLAKCAEEFA EHTNFNKVEL LDFVYSKLSS TIWENREEHD LLKIVRDVRQ TFFRSLEDMG1200
1201 V |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) | |
| View Run | No Comments | Liu X, et al. (2005) | |
| View Run | No Comments | Liu X, et al. (2005) | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
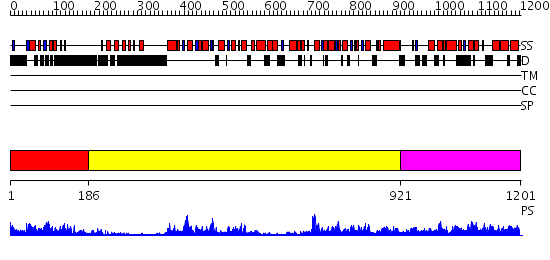
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..185] | 20.30103 | ClpB | |
| 2 | View Details | [186..920] | 120.0 | VCP/p97 | |
| 3 | View Details | [921..1201] | 107.0 | EDTA treated |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)