













| General Information: |
|
| Name(s) found: |
SPBC16D10.08c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 905 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA nucleus [IDA cytoplasm [ISS cytosol [IDA |
| Biological Process: |
chaperone mediated protein folding requiring cofactor
[ISS cellular response to stress [ISS |
| Molecular Function: |
heat shock protein binding
[ISS ATP binding [IEA] chaperone binding [ISS ATPase activity, coupled [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADYPFTDKA AKTLSDAYSI AQSYGHSQLT PIHIAAALLS DSDSNGTTLL RTIVDKAGGD 60
61 GQKFERSVTS RLVRLPAQDP PPEQVTLSPE SAKLLRNAHE LQKTQKDSYI AQDHFIAVFT 120
121 KDDTLKSLLA EAGVTPKAFE FAVNNVRGNK RIDSKNAEEG FDALNKFTVD LTELARNGQL 180
181 DPVIGREDEI RRTIRVLSRR TKNNPVLIGE PGVGKTSIAE GLARRIIDDD VPANLSNCKL 240
241 LSLDVGSLVA GSKFRGEFEE RIKSVLKEVE ESETPIILFV DEMHLLMGAG SGGEGGMDAA 300
301 NLLKPMLARG KLHCIGATTL AEYKKYIEKD AAFERRFQII LVKEPSIEDT ISILRGLKEK 360
361 YEVHHGVTIS DRALVTAAHL ASRYLTSRRL PDSAIDLVDE AAAAVRVTRE SQPEVLDNLE 420
421 RKLRQLRVEI RALEREKDEA SKERLKAARK EAEQVEEETR PIREKYELEK SRGSELQDAK 480
481 RRLDELKAKA EDAERRNDFT LAADLKYYGI PDLQKRIEYL EQQKRKADAE AIANAQPGSE 540
541 PLLIDVVGPD QINEIVARWT GIPVTRLKTT EKERLLNMEK VLSKQVIGQN EAVTAVANAI 600
601 RLSRAGLSDP NQPIASFLFC GPSGTGKTLL TKALASFMFD DENAMIRIDM SEYMEKHSVS 660
661 RLIGAPPGYV GHEAGGQLTE QLRRRPYSVI LFDEIEKAAP EVLTVLLQVL DDGRITSGQG 720
721 QVVDAKNAVI IMTSNLGAEY LTTDNESDDG KIDSTTREMV MNSIRGFFRP EFLNRISSIV 780
781 IFNRLRRVDI RNIVENRILE VQKRLQSNHR SIKIEVSDEA KDLLGSAGYS PAYGARPLNR 840
841 VIQNQVLNPM AVLILNGQLR DKETAHVVVQ NGKIFVKPNH EANANGSADI DMDGIDDDVN 900
901 DEELE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
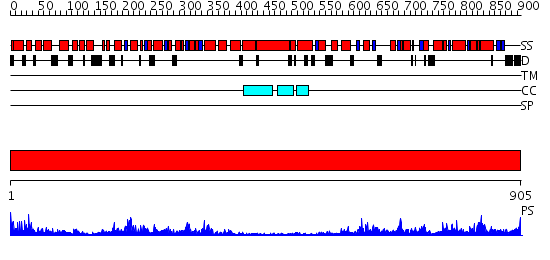
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..905] | 160.0 | Crystal Structure Analysis of ClpB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)